A new method to accurately identify single nucleotide variants using small FFPE breast samples
- PMID: 34117742
- PMCID: PMC8574974
- DOI: 10.1093/bib/bbab221
A new method to accurately identify single nucleotide variants using small FFPE breast samples
Abstract
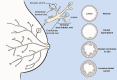
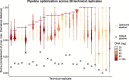
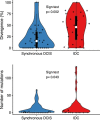
Most tissue collections of neoplasms are composed of formalin-fixed and paraffin-embedded (FFPE) excised tumor samples used for routine diagnostics. DNA sequencing is becoming increasingly important in cancer research and clinical management; however it is difficult to accurately sequence DNA from FFPE samples. We developed and validated a new bioinformatic pipeline to use existing variant-calling strategies to robustly identify somatic single nucleotide variants (SNVs) from whole exome sequencing using small amounts of DNA extracted from archival FFPE samples of breast cancers. We optimized this strategy using 28 pairs of technical replicates. After optimization, the mean similarity between replicates increased 5-fold, reaching 88% (range 0-100%), with a mean of 21.4 SNVs (range 1-68) per sample, representing a markedly superior performance to existing tools. We found that the SNV-identification accuracy declined when there was less than 40 ng of DNA available and that insertion-deletion variant calls are less reliable than single base substitutions. As the first application of the new algorithm, we compared samples of ductal carcinoma in situ of the breast to their adjacent invasive ductal carcinoma samples. We observed an increased number of mutations (paired-samples sign test, P < 0.05), and a higher genetic divergence in the invasive samples (paired-samples sign test, P < 0.01). Our method provides a significant improvement in detecting SNVs in FFPE samples over previous approaches.
Keywords: DCIS; NGS; exome; heterogeneity.
© The Author(s) 2021. Published by Oxford University Press.
Figures

References
-
- McGranahan N, Swanton C. Biological and therapeutic impact of intratumor heterogeneity in cancer evolution. Cancer Cell 2015;27:15–26. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

