MecCog: a knowledge representation framework for genetic disease mechanism
- PMID: 34117883
- PMCID: PMC12161289
- DOI: 10.1093/bioinformatics/btab432
MecCog: a knowledge representation framework for genetic disease mechanism
Abstract
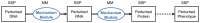
Motivation: Experimental findings on genetic disease mechanisms are scattered throughout the literature and represented in many ways, including unstructured text, cartoons, pathway diagrams and network graphs. Integration and structuring of such mechanistic information greatly enhances its utility.
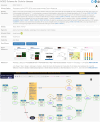
Results: MecCog is a graphical framework for building integrated representations (mechanism schemas) of mechanisms by which a genetic variant causes a disease phenotype. A MecCog mechanism schema displays the propagation of system perturbations across stages of biological organization, using graphical notations to symbolize perturbed entities and activities, hyperlinked evidence tagging, a mechanism ontology and depiction of knowledge gaps, ambiguities and uncertainties. The web platform enables a user to construct, store, publish, browse, query and comment on schemas. MecCog facilitates the identification of potential biomarkers, therapeutic intervention sites and critical future experiments.
Availability and implementation: The MecCog framework is freely available at http://www.meccog.org.
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author(s) 2021. Published by Oxford University Press. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures



Similar articles
-
Harnessing formal concepts of biological mechanism to analyze human disease.PLoS Comput Biol. 2018 Dec 26;14(12):e1006540. doi: 10.1371/journal.pcbi.1006540. eCollection 2018 Dec. PLoS Comput Biol. 2018. PMID: 30586388 Free PMC article.
-
Expediting knowledge acquisition by a web framework for Knowledge Graph Exploration and Visualization (KGEV): case studies on COVID-19 and Human Phenotype Ontology.BMC Med Inform Decis Mak. 2022 Jun 2;22(Suppl 2):147. doi: 10.1186/s12911-022-01848-z. BMC Med Inform Decis Mak. 2022. PMID: 35655307 Free PMC article.
-
A Chado case study: an ontology-based modular schema for representing genome-associated biological information.Bioinformatics. 2007 Jul 1;23(13):i337-46. doi: 10.1093/bioinformatics/btm189. Bioinformatics. 2007. PMID: 17646315
-
Design and development of a linked open data-based health information representation and visualization system: potentials and preliminary evaluation.JMIR Med Inform. 2014 Oct 25;2(2):e31. doi: 10.2196/medinform.3531. JMIR Med Inform. 2014. PMID: 25601195 Free PMC article.
-
Mitochondrial Disease Sequence Data Resource (MSeqDR): a global grass-roots consortium to facilitate deposition, curation, annotation, and integrated analysis of genomic data for the mitochondrial disease clinical and research communities.Mol Genet Metab. 2015 Mar;114(3):388-96. doi: 10.1016/j.ymgme.2014.11.016. Epub 2014 Dec 4. Mol Genet Metab. 2015. PMID: 25542617 Free PMC article. Review.
Cited by
-
AI-powered precision medicine: utilizing genetic risk factor optimization to revolutionize healthcare.NAR Genom Bioinform. 2025 May 5;7(2):lqaf038. doi: 10.1093/nargab/lqaf038. eCollection 2025 Jun. NAR Genom Bioinform. 2025. PMID: 40330081 Free PMC article. Review.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

