Improved SARS-CoV-2 Mpro inhibitors based on feline antiviral drug GC376: Structural enhancements, increased solubility, and micellar studies
- PMID: 34118724
- PMCID: PMC8164773
- DOI: 10.1016/j.ejmech.2021.113584
Improved SARS-CoV-2 Mpro inhibitors based on feline antiviral drug GC376: Structural enhancements, increased solubility, and micellar studies
Abstract
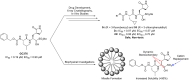
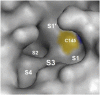
Replication of SARS-CoV-2, the coronavirus causing COVID-19, requires a main protease (Mpro) to cleave viral proteins. Consequently, Mpro is a target for antiviral agents. We and others previously demonstrated that GC376, a bisulfite prodrug with efficacy as an anti-coronaviral agent in animals, is an effective inhibitor of Mpro in SARS-CoV-2. Here, we report structure-activity studies of improved GC376 derivatives with nanomolar affinities and therapeutic indices >200. Crystallographic structures of inhibitor-Mpro complexes reveal that an alternative binding pocket in Mpro, S4, accommodates the P3 position. Alternative binding is induced by polar P3 groups or a nearby methyl. NMR and solubility studies with GC376 show that it exists as a mixture of stereoisomers and forms colloids in aqueous media at higher concentrations, a property not previously reported. Replacement of its Na+ counter ion with choline greatly increases solubility. The physical, biochemical, crystallographic, and cellular data reveal new avenues for Mpro inhibitor design.
Keywords: COVID-19; Crystallography; GC376 analogs; Main protease; Protease inhibitor; Structure-guided design.
Copyright © 2021 Elsevier Masson SAS. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare no competing interests.
Figures


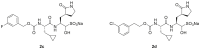
References
-
- COVID-19 map, johns hopkins coronavirus resource center. https://coronavirus.jhu.edu/map.html (n.d.)
-
- A contemporary view of coronavirus transcription | J. Virol., (n.d.). https://jvi.asm.org/content/81/1/20 (accessed February 6, 2021). - PMC - PubMed
-
- Pillaiyar T., Manickam M., Namasivayam V., Hayashi Y., Jung S.-H. An overview of severe acute respiratory syndrome–coronavirus (SARS-CoV) 3CL protease inhibitors: peptidomimetics and small molecule chemotherapy. J. Med. Chem. 2016;59:6595–6628. doi: 10.1021/acs.jmedchem.5b01461. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

