The aging mouse lens transcriptome
- PMID: 34119483
- PMCID: PMC8720273
- DOI: 10.1016/j.exer.2021.108663
The aging mouse lens transcriptome
Abstract
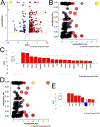
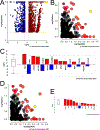
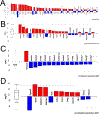
Age is a major risk factor for cataract (ARC). However, the influence of aging on the lens transcriptome is under studied. Lens epithelial (LEC) and fiber cells (LFC) were isolated from young (3 month old) and aged (24 month old) C57BL/6J mice, and the transcriptome elucidated via RNAseq. EdgeR estimated differential gene expression in pairwise contrasts, and Advaita's Ipathway guide and custom R scripts were used to evaluate the potential biological significance of differentially expressed genes (DEGs). This analysis revealed age-dependent decreases in lens differentiation marker expression in both LECs and LFCs, with gamma crystallin transcripts downregulating nearly 50 fold in aged LFCs. The expression of the transcription factors Hsf4 and Maf, which are known to activate lens fiber cell preferred genes, are downregulated, while FoxE3, which represses gamma crystallin expression, is upregulated in aged fibers. Aged LECs upregulate genes controlling the immune response, complement pathways, and cellular stress responses, including glutathione peroxidase 3 (Gpx3). Aged LFCs exhibit broad changes in the expression of genes regulating cell communication, and upregulate genes involved in antigen processing/presentation and cholesterol metabolism, while changes in the expression of mitochondrial respiratory chain genes are consistent with mitochondrial stress, including upregulation of NDufa4l2, which encodes an alternate electron transport chain protein. However, age did not profoundly affect the response of LECs to injury as both young and aged LECs upregulate inflammatory gene signatures at 24 h post injury to similar extents. These RNAseq profiles provide a rich data set that can be mined to understand the genetic regulation of lens aging and how this impinges on the pathophysiology of age related cataract.
Keywords: Aging; Cholestero; Complement pathways; Lens; Metabolism; Posterior capsular opacification; Senescence; Transcriptome.
Copyright © 2021. Published by Elsevier Ltd.
Figures


References
-
- Agrawal SA, Anand D, Siddam AD, Kakrana A, Dash S, Scheiblin DA, Dang CA, Terrell AM, Waters SM, Singh A, Motohashi H, Yamamoto M, Lachke SA, 2015. Compound mouse mutants of bZIP transcription factors Mafg and Mafk reveal a regulatory network of non-crystallin genes associated with cataract. Hum Genet 134, 717–735. - PMC - PubMed
-
- Ahsan S, Draghici S, 2017. Identifying Significantly Impacted Pathways and Putative Mechanisms with iPathwayGuide. Curr Protoc Bioinformatics 57, 7 15 11–17 15 30. - PubMed
-
- Aleo MD, Doshna CM, Baltrukonis D, Fortner JH, Drupa CA, Navetta KA, Fritz CA, Potter DM, Verdugo ME, Beierschmitt WP, 2019. Lens cholesterol biosynthesis inhibition: A common mechanism of cataract formation in laboratory animals by pharmaceutical products. J Appl Toxicol 39, 1348–1361. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

