A small ribosome-associated ncRNA globally inhibits translation by restricting ribosome dynamics
- PMID: 34121604
- PMCID: PMC8632108
- DOI: 10.1080/15476286.2021.1935573
A small ribosome-associated ncRNA globally inhibits translation by restricting ribosome dynamics
Abstract
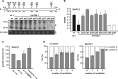
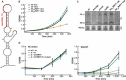
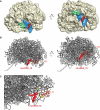
Ribosome-associated non-coding RNAs (rancRNAs) have been recognized as an emerging class of regulatory molecules capable of fine-tuning translation in all domains of life. RancRNAs are ideally suited for allowing a swift response to changing environments and are therefore considered pivotal during the first wave of stress adaptation. Previously, we identified an mRNA-derived 18 nucleotides long rancRNA (rancRNA_18) in Saccharomyces cerevisiae that rapidly downregulates protein synthesis during hyperosmotic stress. However, the molecular mechanism of action remained enigmatic. Here, we combine biochemical, genetic, transcriptome-wide and structural evidence, thus revealing rancRNA_18 as global translation inhibitor by targeting the E-site region of the large ribosomal subunit. Ribosomes carrying rancRNA_18 possess decreased affinity for A-site tRNA and impaired structural dynamics. Cumulatively, these discoveries reveal the mode of action of a rancRNA involved in modulating protein biosynthesis at a thus far unequalled precision.
Keywords: Non-coding rna; l1 stalk; rancRNA; ribosome; translation control.
Figures



References
-
- Lane N, Martin W.. The energetics of genome complexity. Nature. 2010;467(7318):929. - PubMed
-
- Gonskikh Y, Polacek N.. Alterations of the translation apparatus during aging and stress response. Mech Ageing Dev. 2017;168:30–36. - PubMed
-
- Sherman MY, Qian SB. Less is more: improving proteostasis by translation slow down. Trends Biochem Sci. 2013;38(12):585–591. - PubMed
-
- Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev. 2010;11(9):597–610. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
