Aux/IAA and ARF Gene Families in Salix suchowensis: Identification, Evolution, and Dynamic Transcriptome Profiling During the Plant Growth Process
- PMID: 34122487
- PMCID: PMC8188177
- DOI: 10.3389/fpls.2021.666310
Aux/IAA and ARF Gene Families in Salix suchowensis: Identification, Evolution, and Dynamic Transcriptome Profiling During the Plant Growth Process
Abstract
The phytohormone auxin plays a pivotal role in the regulation of plant growth and development, including vascular differentiation and tree growth. The auxin/indole-3-acetic acid (Aux/IAA) and auxin response transcription factor (ARF) genes are key components of plant auxin signaling. To gain more insight into the regulation and functional features of Aux/IAA and ARF genes during these processes, we identified 38 AUX/IAA and 34 ARF genes in the genome of Salix suchowensis and characterized their gene structures, conserved domains, and encoded amino acid compositions. Phylogenetic analysis of some typical land plants showed that the Aux/IAA and ARF genes of Salicaceae originated from a common ancestor and were significantly amplified by the ancestral eudicot hexaploidization event and the "salicoid" duplication that occurred before the divergence of poplar and willow. By analyzing dynamic transcriptome profiling data, some Aux/IAA and ARF genes were found to be involved in the regulation of plant growth, especially in the initial plant growth process. Additionally, we found that the expression of several miR160/miR167-ARFs was in agreement with canonical miRNA-ARF interactions, suggesting that miRNAs were possibly involved in the regulation of the auxin signaling pathway and the plant growth process. In summary, this study comprehensively analyzed the sequence features, origin, and expansion of Aux/IAA and ARF genes, and the results provide useful information for further studies on the functional involvement of auxin signaling genes in the plant growth process.
Keywords: Aux/IAA and ARF gene families; auxin signaling; dynamic transcriptome profiling; plant growth process; polyploidization events.
Copyright © 2021 Wei, Chen, Hou, Yang and Yin.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
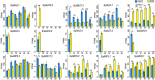
Figures



References
-
- Andersson-Gunnerås S., Mellerowicz E. J., Love J., Segerman B., Ohmiya Y., Coutinho P. M., et al. (2006). Biosynthesis of cellulose-enriched tension wood in populus: global analysis of transcripts and metabolites identifies biochemical and developmental regulators in secondary wall biosynthesis. Plant J. 45 144–165. 10.1111/j.1365-313X.2005.02584.x - DOI - PubMed
-
- Berleth T., Jurgens G. (1993). The role of the monopteros gene in organising the basal body region of the Arabidopsis embryo. Development 118:575.
-
- Bhalerao R. P., Fischer U. (2014). Auxin gradients across wood-instructive or incidental? Physiol. Plant. 151 43–51. - PubMed
LinkOut - more resources
Full Text Sources

