A highly stable RNA aptamer probe for the retinoblastoma protein in live cells
- PMID: 34122904
- PMCID: PMC8159449
- DOI: 10.1039/d0sc01613f
A highly stable RNA aptamer probe for the retinoblastoma protein in live cells
Abstract
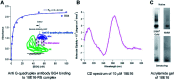
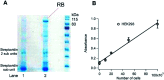
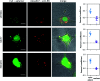
Although RNA aptamers can show comparable or better specificity and affinity to antibodies and have the advantage of being able to access different live cell compartments, they are often much less stable in vivo. We report here the first aptamer that binds human retinoblastoma protein (RB) and is stable in live cells. RB is both a key protein in cell cycle control and also a tumour suppressor. The aptamer was selected from an RNA library against a unique 12-residue helical peptide derived from RB rather than the whole protein molecule. It binds RB with high affinity (K d = 5.1 ± 0.1 nM) and is a putative RNA G-quadruplex structure formed by an 18-nucleotide sequence (18E16 - GGA GGG UGG AGG GAA GGG), which may account for its high stability. Confocal fluorescence microscopy of live cells transfected with the aptamer shows it is stable intracellularly and efficient in entering the nucleus where an analogous antibody was inaccessible. The findings demonstrate this aptamer is an advanced probe for RB in live cell applications.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare.
Figures

Similar articles
-
Gold nanoparticle-streptavidin conjugates for rapid and efficient screening of aptamer function in lateral flow sensors using novel CD4-binding aptamers identified through Crossover-SELEX.Analyst. 2020 Aug 7;145(15):5180-5193. doi: 10.1039/d0an00634c. Epub 2020 Jun 22. Analyst. 2020. PMID: 32567629
-
Selection of aptamers against live bacterial cells.Anal Chem. 2008 Oct 15;80(20):7812-9. doi: 10.1021/ac801272s. Epub 2008 Sep 20. Anal Chem. 2008. PMID: 18803393
-
Aptamer Binding Assay for the E Antigen of Hepatitis B Using Modified Aptamers with G-Quadruplex Structures.Anal Chem. 2020 May 5;92(9):6495-6501. doi: 10.1021/acs.analchem.9b05740. Epub 2020 Apr 16. Anal Chem. 2020. PMID: 32250595
-
An Aptamer-Based Probe for Molecular Subtyping of Breast Cancer.Theranostics. 2018 Nov 10;8(20):5772-5783. doi: 10.7150/thno.28949. eCollection 2018. Theranostics. 2018. PMID: 30555580 Free PMC article.
-
A label-free fluorescence assay for thrombin based on aptamer exonuclease protection and exonuclease III-assisted recycling amplification-responsive cascade zinc(II)-protoporphyrin IX/G-quadruplex supramolecular fluorescent labels.Analyst. 2014 May 21;139(10):2583-8. doi: 10.1039/c3an02336b. Analyst. 2014. PMID: 24707508
Cited by
-
Retinoblastoma: A review of the molecular basis of tumor development and its clinical correlation in shaping future targeted treatment strategies.Indian J Ophthalmol. 2023 Jul;71(7):2662-2676. doi: 10.4103/IJO.IJO_3172_22. Indian J Ophthalmol. 2023. PMID: 37417104 Free PMC article. Review.
-
Development of high affinity broadly reactive aptamers for spike protein of multiple SARS-CoV-2 variants.RSC Adv. 2023 May 19;13(22):15322-15326. doi: 10.1039/d3ra01382k. eCollection 2023 May 15. RSC Adv. 2023. PMID: 37213341 Free PMC article.
-
Subcellular Transcriptomics and Proteomics: A Comparative Methods Review.Mol Cell Proteomics. 2022 Feb;21(2):100186. doi: 10.1016/j.mcpro.2021.100186. Epub 2021 Dec 16. Mol Cell Proteomics. 2022. PMID: 34922010 Free PMC article. Review.
-
Analyzing aptamer structure and interactions: in silico modelling and instrumental methods.Biophys Rev. 2024 Nov 20;16(6):685-700. doi: 10.1007/s12551-024-01252-z. eCollection 2024 Dec. Biophys Rev. 2024. PMID: 39830127 Review.
References
LinkOut - more resources
Full Text Sources
Miscellaneous

