A straightforward approach to antibodies recognising cancer specific glycopeptidic neoepitopes
- PMID: 34122956
- PMCID: PMC8159228
- DOI: 10.1039/d0sc00317d
A straightforward approach to antibodies recognising cancer specific glycopeptidic neoepitopes
Erratum in
-
Correction: A straightforward approach to antibodies recognising cancer specific glycopeptidic neoepitopes.Chem Sci. 2020 Nov 18;11(46):12588-12589. doi: 10.1039/d0sc90254c. Chem Sci. 2020. PMID: 34125114 Free PMC article.
Abstract
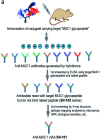
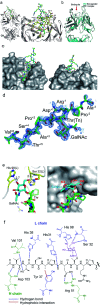
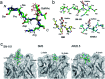
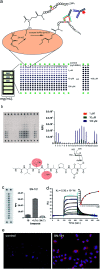
Aberrantly truncated immature O-glycosylation in proteins occurs in essentially all types of epithelial cancer cells, which was demonstrated to be a common feature of most adenocarcinomas and strongly associated with cancer proliferation and metastasis. Although extensive efforts have been made toward the development of anticancer antibodies targeting MUC1, one of the most studied mucins having cancer-relevant immature O-glycans, no anti-MUC1 antibody recognises carbohydrates and the proximal MUC1 peptide region, concurrently. Here we present a general strategy that allows for the creation of antibodies interacting specifically with glycopeptidic neoepitopes by using homogeneous synthetic MUC1 glycopeptides designed for the streamlined process of immunization, antibody screening, three-dimensional structure analysis, epitope mapping and biochemical analysis. The X-ray crystal structure of the anti-MUC1 monoclonal antibody SN-101 complexed with the antigenic glycopeptide provides for the first time evidence that SN-101 recognises specifically the essential epitope by forming multiple hydrogen bonds both with the proximal peptide and GalNAc linked to the threonine residue, concurrently. Remarkably, the structure of the MUC1 glycopeptide in complex with SN-101 is identical to its solution NMR structure, an extended conformation induced by site-specific glycosylation. We demonstrate that this method accelerates dramatically the development of a new class of designated antibodies targeting a variety of "dynamic neoepitopes" elaborated by disease-specific O-glycosylation in the immunodominant mucin domains and mucin-like sequences found in intrinsically disordered regions of many proteins.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare.
Figures
References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

