Target identification among known drugs by deep learning from heterogeneous networks
- PMID: 34123272
- PMCID: PMC8150105
- DOI: 10.1039/c9sc04336e
Target identification among known drugs by deep learning from heterogeneous networks
Abstract
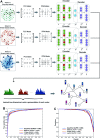
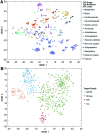
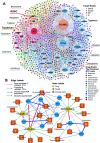
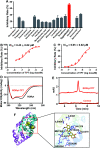
Without foreknowledge of the complete drug target information, development of promising and affordable approaches for effective treatment of human diseases is challenging. Here, we develop deepDTnet, a deep learning methodology for new target identification and drug repurposing in a heterogeneous drug-gene-disease network embedding 15 types of chemical, genomic, phenotypic, and cellular network profiles. Trained on 732 U.S. Food and Drug Administration-approved small molecule drugs, deepDTnet shows high accuracy (the area under the receiver operating characteristic curve = 0.963) in identifying novel molecular targets for known drugs, outperforming previously published state-of-the-art methodologies. We then experimentally validate that deepDTnet-predicted topotecan (an approved topoisomerase inhibitor) is a new, direct inhibitor (IC50 = 0.43 μM) of human retinoic-acid-receptor-related orphan receptor-gamma t (ROR-γt). Furthermore, by specifically targeting ROR-γt, topotecan reveals a potential therapeutic effect in a mouse model of multiple sclerosis. In summary, deepDTnet offers a powerful network-based deep learning methodology for target identification to accelerate drug repurposing and minimize the translational gap in drug development.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
The conflict of interest is managed by the Conflict of Interest Committee of Cleveland Clinic in accordance with its conflict of interest policies. JL is a co-founder of Scipher Medicine, Inc. FC is an inventor on a pending US patent application for this network-based, deep learning technology. The other authors declare no competing interests.
Figures


References
-
- Avorn J. Engl N. J. Med. 2015;372:1877–1879. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

