Chemical tagging for sensitive determination of uridine modifications in RNA
- PMID: 34123281
- PMCID: PMC8148390
- DOI: 10.1039/c9sc05094a
Chemical tagging for sensitive determination of uridine modifications in RNA
Abstract
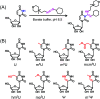
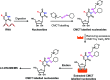
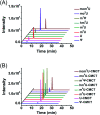
The discovery of dynamic and reversible modifications in messenger RNA (mRNA) is opening new directions in RNA modification-mediated regulation of biological processes. Methylation is the most prevalent modification occurring in mRNA and the methyl group is mainly decorated in the adenine, cytosine, and guanine base or in the 2'-hydroxyl group of ribose. However, methylation of the uracil base (5-methyluridine, m5U) has not been discovered in mRNA of eukaryotes. In the current study, we established a method of N-cyclohexyl-N'-β-(4-methylmorpholinium) ethylcarbodiimide p-toluenesulfonate (CMCT) labelling coupled with liquid chromatography-electrospray ionization-mass spectrometry (LC-ESI-MS/MS) analysis for the sensitive determination of uridine modifications in RNA. Our results demonstrated that the detection sensitivities of uridine modifications in RNA increased up to 1408 fold upon CMCT labelling. Using the developed method, we identified the distinct existence of m5U in mRNA of various mammalian cells and tissues. In addition, the stable isotope tracing monitored by mass spectrometry revealed that the methyl group of m5U originated from S-adenosyl-l-methionine (SAM). Our study expanded the list of modifications occurring in mRNA of mammals. Future work on transcriptome-wide mapping of m5U will further uncover the functional roles of m5U in mRNA of mammals.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
The authors declare no competing financial interest.
Figures







References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

