Therapeutic Targeting of Notch Signaling: From Cancer to Inflammatory Disorders
- PMID: 34124039
- PMCID: PMC8194077
- DOI: 10.3389/fcell.2021.649205
Therapeutic Targeting of Notch Signaling: From Cancer to Inflammatory Disorders
Abstract
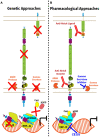
Over the past two decades, the Notch signaling pathway has been investigated as a therapeutic target for the treatment of cancers, and more recently in the context of immune and inflammatory disorders. Notch is an evolutionary conserved pathway found in all metazoans that is critical for proper embryonic development and for the postnatal maintenance of selected tissues. Through cell-to-cell contacts, Notch orchestrates cell fate decisions and differentiation in non-hematopoietic and hematopoietic cell types, regulates immune cell development, and is integral to shaping the amplitude as well as the quality of different types of immune responses. Depriving some cancer types of Notch signals has been shown in preclinical studies to stunt tumor growth, consistent with an oncogenic function of Notch signaling. In addition, therapeutically antagonizing Notch signals showed preclinical potential to prevent or reverse inflammatory disorders, including autoimmune diseases, allergic inflammation and immune complications of life-saving procedures such allogeneic bone marrow and solid organ transplantation (graft-versus-host disease and graft rejection). In this review, we discuss some of these unique approaches, along with the successes and challenges encountered so far to target Notch signaling in preclinical and early clinical studies. Our goal is to emphasize lessons learned to provide guidance about emerging strategies of Notch-based therapeutics that could be deployed safely and efficiently in patients with immune and inflammatory disorders.
Keywords: Notch; Notch ligands; cancer; immune system; inflammation.
Copyright © 2021 Allen and Maillard.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

