Spc1 regulates the signal peptidase-mediated processing of membrane proteins
- PMID: 34125229
- PMCID: PMC8277137
- DOI: 10.1242/jcs.258936
Spc1 regulates the signal peptidase-mediated processing of membrane proteins
Abstract
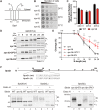
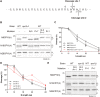
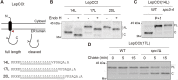
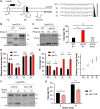
Signal peptidase (SPase) cleaves the signal sequences (SSs) of secretory precursors. It contains an evolutionarily conserved membrane protein subunit, Spc1, that is dispensable for the catalytic activity of SPase and whose role remains unknown. In this study, we investigated the function of yeast Spc1. First, we set up an in vivo SPase cleavage assay using variants of the secretory protein carboxypeptidase Y (CPY) with SSs modified in the N-terminal and hydrophobic core regions. When comparing the SS cleavage efficiencies of these variants in cells with or without Spc1, we found that signal-anchored sequences became more susceptible to cleavage by SPase without Spc1. Furthermore, SPase-mediated processing of model membrane proteins was enhanced in the absence of Spc1 and was reduced upon overexpression of Spc1. Spc1 co-immunoprecipitated with proteins carrying uncleaved signal-anchored or transmembrane (TM) segments. Taken together, these results suggest that Spc1 protects TM segments from SPase action, thereby sharpening SPase substrate selection and acting as a negative regulator of the SPase-mediated processing of membrane proteins.
Keywords: SPCS1; Signal peptidase; Signal sequence; Spc1; Transmembrane.
© 2021. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures

References
-
- Barkocy-Gallagher, G. A. and Bassford, P. J.Jr. (1992). Synthesis of precursor maltose-binding protein with proline in the +1 position of the cleavage site interferes with the activity of Escherichia coli signal peptidase I in vivo. J. Biol. Chem. 267, 1231-1238. 10.1016/S0021-9258(18)48419-0 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

