Free energy landscape of RNA binding dynamics in start codon recognition by eukaryotic ribosomal pre-initiation complex
- PMID: 34125830
- PMCID: PMC8224888
- DOI: 10.1371/journal.pcbi.1009068
Free energy landscape of RNA binding dynamics in start codon recognition by eukaryotic ribosomal pre-initiation complex
Abstract
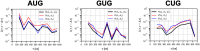
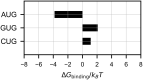
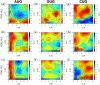
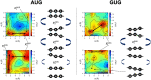
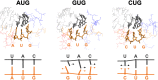
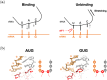
Specific interaction between the start codon, 5'-AUG-3', and the anticodon, 5'-CAU-3', ensures accurate initiation of translation. Recent studies show that several near-cognate start codons (e.g. GUG and CUG) can play a role in initiating translation in eukaryotes. However, the mechanism allowing initiation through mismatched base-pairs at the ribosomal decoding site is still unclear at an atomic level. In this work, we propose an extended simulation-based method to evaluate free energy profiles, through computing the distance between each base-pair of the triplet interactions involved in recognition of start codons in eukaryotic translation pre-initiation complex. Our method provides not only the free energy penalty for mismatched start codons relative to the AUG start codon, but also the preferred pathways of transitions between bound and unbound states, which has not been described by previous studies. To verify the method, the binding dynamics of cognate (AUG) and near-cognate start codons (CUG and GUG) were simulated. Evaluated free energy profiles agree with experimentally observed changes in initiation frequencies from respective codons. This work proposes for the first time how a G:U mismatch at the first position of codon (GUG)-anticodon base-pairs destabilizes the accommodation in the initiating eukaryotic ribosome and how initiation at a CUG codon is nearly as strong as, or sometimes stronger than, that at a GUG codon. Our method is expected to be applied to study the affinity changes for various mismatched base-pairs.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

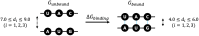
References
-
- Asano K. Translational control. In: Encyclopedia of Systems Biology. Springer; New York; 2013. p. 2278–2282.
-
- Asano K, Ito K. Translation elongation. In: Encyclopedia of Systems Biology. Springer; New York; 2013. p. 2259–2263.
-
- Hinnebusch AG, Dever TE, Asano K. Mechanism of translation initiation in the yeast Saccharomyces cerevisiae. Cold Spring Harbor Monograph Series. 2007;48:225–268.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

