A new assay capturing chromosome fusions shows a protection trade-off at telomeres and NHEJ vulnerability to low-density ionizing radiation
- PMID: 34125900
- PMCID: PMC8266670
- DOI: 10.1093/nar/gkab502
A new assay capturing chromosome fusions shows a protection trade-off at telomeres and NHEJ vulnerability to low-density ionizing radiation
Abstract
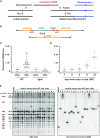
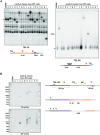
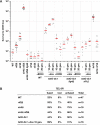
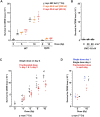
Chromosome fusions threaten genome integrity and promote cancer by engaging catastrophic mutational processes, namely chromosome breakage-fusion-bridge cycles and chromothripsis. Chromosome fusions are frequent in cells incurring telomere dysfunctions or those exposed to DNA breakage. Their occurrence and therefore their contribution to genome instability in unchallenged cells is unknown. To address this issue, we constructed a genetic assay able to capture and quantify rare chromosome fusions in budding yeast. This chromosome fusion capture (CFC) assay relies on the controlled inactivation of one centromere to rescue unstable dicentric chromosome fusions. It is sensitive enough to quantify the basal rate of end-to-end chromosome fusions occurring in wild-type cells. These fusions depend on canonical nonhomologous end joining (NHEJ). Our results show that chromosome end protection results from a trade-off at telomeres between positive effectors (Rif2, Sir4, telomerase) and a negative effector partially antagonizing them (Rif1). The CFC assay also captures NHEJ-dependent chromosome fusions induced by ionizing radiation. It provides evidence for chromosomal rearrangements stemming from a single photon-matter interaction.
© The Author(s) 2021. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



References
-
- Lee K., Zhang Y., Lee S.E.. Saccharomyces cerevisiae ATM orthologue suppresses break-induced chromosome translocations. Nature. 2008; 454:543–546. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

