Mechanisms of Transforming DNA Uptake to the Periplasm of Bacillus subtilis
- PMID: 34126763
- PMCID: PMC8262900
- DOI: 10.1128/mBio.01061-21
Mechanisms of Transforming DNA Uptake to the Periplasm of Bacillus subtilis
Abstract
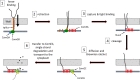
We demonstrate here that the acquisition of DNase resistance by transforming DNA, often assumed to indicate transport to the cytoplasm, reflects uptake to the periplasm, requiring a reevaluation of conclusions about the roles of several proteins in transformation. The new evidence suggests that the transformation pilus is needed for DNA binding to the cell surface near the cell poles and for the initiation of uptake. The cellular distribution of the membrane-anchored ComEA of Bacillus subtilis does not dramatically change during DNA uptake as does the unanchored ComEA of Vibrio and Neisseria. Instead, our evidence suggests that ComEA stabilizes the attachment of transforming DNA at localized regions in the periplasm and then mediates uptake, probably by a Brownian ratchet mechanism. Following that, the DNA is transferred to periplasmic portions of the channel protein ComEC, which plays a previously unsuspected role in uptake to the periplasm. We show that the transformation endonuclease NucA also facilitates uptake to the periplasm and that the previously demonstrated role of ComFA in the acquisition of DNase resistance derives from the instability of ComGA when ComFA is deleted. These results prompt a new understanding of the early stages of DNA uptake for transformation. IMPORTANCE Transformation is a widely distributed mechanism of bacterial horizontal gene transfer that plays a role in the spread of antibiotic resistance and virulence genes and more generally in evolution. Although transformation was discovered nearly a century ago and most, if not all the proteins required have been identified in several bacterial species, much remains poorly understood about the molecular mechanism of DNA uptake. This study uses epifluorescence microscopy to investigate the passage of labeled DNA into the compartment between the cell wall and the cell membrane of Bacillus subtilis, a necessary early step in transformation. The roles of individual proteins in this process are identified, and their modes of action are clarified.
Keywords: Bacillus subtilis; ComEA; ComEC; DNA uptake; periplasm; transformation.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
