Identification of novel hub genes associated with gastric cancer using integrated bioinformatics analysis
- PMID: 34126961
- PMCID: PMC8201699
- DOI: 10.1186/s12885-021-08358-7
Identification of novel hub genes associated with gastric cancer using integrated bioinformatics analysis
Abstract
Background: Gastric cancer (GC) is one of the most common solid malignant tumors worldwide with a high-recurrence-rate. Identifying the molecular signatures and specific biomarkers of GC might provide novel clues for GC prognosis and targeted therapy.
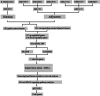
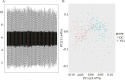
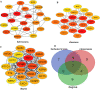
Methods: Gene expression profiles were obtained from the ArrayExpress and Gene Expression Omnibus database. Differentially expressed genes (DEGs) were picked out by R software. The hub genes were screened by cytohubba plugin. Their prognostic values were assessed by Kaplan-Meier survival analyses and the gene expression profiling interactive analysis (GEPIA). Finally, qRT-PCR in GC tissue samples was established to validate these DEGs.
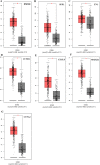
Results: Total of 295 DEGs were identified between GC and their corresponding normal adjacent tissue samples in E-MTAB-1440, GSE79973, GSE19826, GSE13911, GSE27342, GSE33335 and GSE56807 datasets, including 117 up-regulated and 178 down-regulated genes. Among them, 7 vital upregulated genes (HMMR, SPP1, FN1, CCNB1, CXCL8, MAD2L1 and CCNA2) were selected. Most of them had a significantly worse prognosis except SPP1. Using qRT-PCR, we validated that their transcriptions in our GC tumor tissue were upregulated except SPP1 and FN1, which correlated with tumor relapse and predicts poorer prognosis in GC patients.
Conclusions: We have identified 5 upregulated DEGs (HMMR, CCNB1, CXCL8, MAD2L1, and CCNA2) in GC patients with poor prognosis using integrated bioinformatical methods, which could be potential biomarkers and therapeutic targets for GC treatment.
Keywords: Bioinformatics analysis; Differentially expressed genes; Gastric cancer.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

