RNAi of a Putative Grapevine Susceptibility Gene as a Possible Downy Mildew Control Strategy
- PMID: 34127927
- PMCID: PMC8196239
- DOI: 10.3389/fpls.2021.667319
RNAi of a Putative Grapevine Susceptibility Gene as a Possible Downy Mildew Control Strategy
Abstract
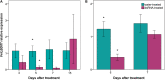
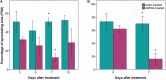
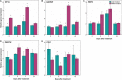
Downy mildew, caused by the oomycete Plasmopara viticola, is one of the diseases causing the most severe economic losses to grapevine (Vitis vinifera) production. To date, the application of fungicides is the most efficient method to control the pathogen and the implementation of novel and sustainable disease control methods is a major challenge. RNA interference (RNAi) represents a novel biotechnological tool with a great potential for controlling fungal pathogens. Recently, a candidate susceptibility gene (VviLBDIf7) to downy mildew has been identified in V. vinifera. In this work, the efficacy of RNAi triggered by exogenous double-stranded RNA (dsRNA) in controlling P. viticola infections has been assessed in a highly susceptible grapevine cultivar (Pinot noir) by knocking down VviLBDIf7 gene. The effects of dsRNA treatment on this target gene were assessed by evaluating gene expression, disease severity, and development of vegetative and reproductive structures of P. viticola in the leaf tissues. Furthermore, the effects of dsRNA treatment on off-target (EF1α, GAPDH, PEPC, and PEPCK) and jasmonic acid metabolism (COI1) genes have been evaluated. Exogenous application of dsRNA led to significant reductions both in VviLBDIf7 gene expression, 5 days after the treatment, and in the disease severity when artificial inoculation was carried out 7 days after dsRNA treatments. The pathogen showed clear alterations to both vegetative (hyphae and haustoria) and reproductive structures (sporangiophores) that resulted in stunted growth and reduced sporulation. Treatment with dsRNA showed signatures of systemic activity and no deleterious off-target effects. These results demonstrated the potential of RNAi for silencing susceptibility factors in grapevine as a sustainable strategy for pathogen control, underlying the possibility to adopt this promising biotechnological tool in disease management strategies.
Keywords: Vitis vinifera; disease resistance; dsRNA; gene silencing; obligate parasite; susceptibility gene.
Copyright © 2021 Marcianò, Ricciardi, Marone Fassolo, Passera, Bianco, Failla, Casati, Maddalena, De Lorenzis and Toffolatti.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Bachewich C., Heath I. B. (1999). Cytoplasmic migrations and vacuolation are associated with growth recovery in hyphae of Saprolegnia, and are dependent on the cytoskeleton. Mycol. Res. 103 849–858.
-
- Biedenkopf D., Will T., Knauer T., Jelonek L., Furch A. C. U., Busche T., et al. (2020). Systemic spreading of exogenous applied RNA biopesticides in the crop plant Hordeum vulgare. ExRNA 2:12.
LinkOut - more resources
Full Text Sources
Research Materials

