Identification of a prognostic metabolic gene signature in diffuse large B-cell lymphoma
- PMID: 34128320
- PMCID: PMC8278125
- DOI: 10.1111/jcmm.16720
Identification of a prognostic metabolic gene signature in diffuse large B-cell lymphoma
Abstract
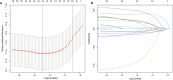
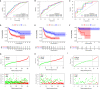
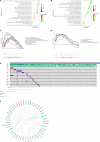
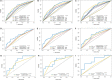
Diffuse large B-cell lymphoma (DLBCL) is a clinically diverse disease. Given the numerous genetic mutations and variations associated with it, a prognostic gene signature that can be related to the overall survival (OS) is a clinical implication. We used the mRNA expression profiles and clinicopathological data of patients with DLBCL from the Gene Expression Omnibus (GEO) database to identify a metabolism-related gene signature. Using LASSO regression analysis, a novel 13-metabolic gene signature was identified to evaluate prognosis. The information gathered was used to construct the nomogram model to improve risk stratification and quantify risk factors for individual patients. We performed gene set enrichment analysis to identify the enriched signalling axes to further understand the underlying biological pathways. The receiver operating characteristic (ROC) curve revealed a satisfactory performance in the training cohorts. The model also showed clinical benefit when compared to the standard prognostic factors (P < .05) in validation cohorts. This study aimed to combine metabolic dysregulation with clinical features of patients with DLBCL to generate a prognostic model that might not only indicate the value of the metabolic microenvironment for prognostic stratification but also improve the decision-making during individual therapy.
Keywords: clinical prognostic model; diffuse large B-cell lymphoma; gene signature; metabolism.
© 2021 The Authors. Journal of Cellular and Molecular Medicine published by Foundation for Cellular and Molecular Medicine and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest in this work.
Figures



References
-
- Lauren RT, DeSantis CE, Cerhan JR, et al. 2016 US lymphoid malignancy statistics by World Health Organization subtypes. CA Cancer J Clin. 2016;6(66):443‐459. - PubMed
-
- Swerdlow S. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. 2017. Lyon, France: World Health Organization;
-
- Coiffier B, Lepage E, Brière J, et al. CHOP chemotherapy plus rituximab compared WITH CHOP alone in elderly patients with diffuse large‐B‐Cell Lymphoma. N Engl J Med. 2002;346:235‐242. - PubMed
-
- Mareschal S, Ruminy P, Bagacean C, et al. Accurate classification of germinal center B‐Cell‐Like/Activated B‐Cell‐Like diffuse large B‐Cell lymphoma using a simple and rapid reverse transcriptase‐multiplex ligation‐dependent probe amplification assay: a CALYM Study. Journal of Molecular Diagnostics. 2015;15(S1525–1578):273‐283. - PubMed
-
- International Non‐Hodgkin's Lymphoma Prognostic Factors Project .A predictive model for aggressive non‐Hodgkin’s lymphoma. The International Non‐Hodgkin's Lymphoma Prognostic Factors Project. N Engl J Med. 1993;14(329):987‐994. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

