Immune infiltration phenotypes of prostate adenocarcinoma and their clinical implications
- PMID: 34128342
- PMCID: PMC8335836
- DOI: 10.1002/cam4.4063
Immune infiltration phenotypes of prostate adenocarcinoma and their clinical implications
Abstract
Background: Tumor-infiltrating immune cells participate in the initiation and progression of prostate adenocarcinoma (PRAD). However, it is not fully known how immune infiltration affects the development of PRAD and its clinical presentation.
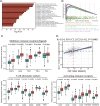
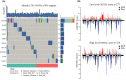
Methods: Herein, we investigated the immune infiltration phenotypes in PRAD based on transcriptome profiles, methylation profiles, somatic mutation, and copy number variations. We also developed an immune prognostic model (IPM) to identify unfavorable prognosis. To verify this model, immunohistochemistry staining was performed on a cohort of PRAD samples. Moreover, we constructed a nomogram to assess the survival of PRAD incorporating immune infiltration and other clinical features.
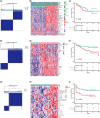
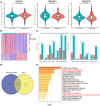
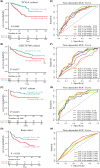
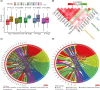
Results: We categorized PRAD patients into high and low-level clusters based on immune infiltration phenotypes. The patients in the high-level clusters had worse survival than their low-level counterparts. Gene set enrichment analysis indicated that both anti- and pro-tumor terms were enriched in high-level cluster. Moreover, we identified a positive correlation between anti- and pro-tumor immune cells in PRAD microenvironment. Notably, Somatic mutation analysis showed patients in high-level cluster had a higher somatic mutation burden of KMT2D, HSPA8, CHD7, and MAP1A. In addition, we developed an IPM with robust predictive ability. The model can distinguish high-risk PRAD patients with poor prognosis from low-risk PRAD patients in both training and another three independent validation datasets. Besides, we constructed a nomogram incorporating Gleason score, pathological T stage, and IPM for the prognosis prediction of PRAD patients, which displayed robust predictive ability and might contribute to clinical practice.
Conclusion: Our work illustrated the immune infiltration phenotypes strongly related to the poor prognosis of PRAD patients, and highlighted the potential of the IPM to identify unfavorable tumor features.
Keywords: biological behaviors; genomic patterns; immune infiltration phenotypes; prognostic signature; prostate adenocarcinoma.
© 2021 The Authors. Cancer Medicine published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures

References
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7‐30. - PubMed
-
- Attard G, Parker C, Eeles RA, et al. Prostate cancer. Lancet (London, England). 2016;387(10013):70‐82. - PubMed
-
- Vinay DS, Ryan EP, Pawelec G, et al. Immune evasion in cancer: Mechanistic basis and therapeutic strategies. Semin Cancer Biol. 2015;S185‐S198. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 81702542 81772742 81972578 82072847/National Natural Science Foundation of China
- 19XD1402300/Science and Technology Commission of Shanghai Municipality
- 2019LJ11/Shanghai Municipal Health Commission
- YG2019GD02/Shanghai Jiao Tong University Medical Engineering Cross Fund
- 2018WS275/Shandong Medical and Health Science and Technology Development Plan Project
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

