The cryptic gonadotropin-releasing hormone neuronal system of human basal ganglia
- PMID: 34128468
- PMCID: PMC8245125
- DOI: 10.7554/eLife.67714
The cryptic gonadotropin-releasing hormone neuronal system of human basal ganglia
Abstract
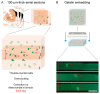
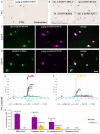
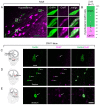
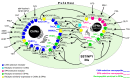
Human reproduction is controlled by ~2000 hypothalamic gonadotropin-releasing hormone (GnRH) neurons. Here, we report the discovery and characterization of additional ~150,000-200,000 GnRH-synthesizing cells in the human basal ganglia and basal forebrain. Nearly all extrahypothalamic GnRH neurons expressed the cholinergic marker enzyme choline acetyltransferase. Similarly, hypothalamic GnRH neurons were also cholinergic both in embryonic and adult human brains. Whole-transcriptome analysis of cholinergic interneurons and medium spiny projection neurons laser-microdissected from the human putamen showed selective expression of GNRH1 and GNRHR1 autoreceptors in the cholinergic cell population and uncovered the detailed transcriptome profile and molecular connectome of these two cell types. Higher-order non-reproductive functions regulated by GnRH under physiological conditions in the human basal ganglia and basal forebrain require clarification. The role and changes of GnRH/GnRHR1 signaling in neurodegenerative disorders affecting cholinergic neurocircuitries, including Parkinson's and Alzheimer's diseases, need to be explored.
Keywords: GnRH; RNA-sequencing; cell biology; cholinergic interneurons; human; human transcriptomics; neuropeptides; neuroscience; striatum.
© 2021, Skrapits et al.
Conflict of interest statement
KS, MS, IF, BG, ST, ÉR, VV, CV, GR, AM, NS, SP, BT, FE, GS, MC, CA, LC, VP, PG, EH No competing interests declared
Figures


References
-
- Bonsi P, Martella G, Cuomo D, Platania P, Sciamanna G, Bernardi G, Wess J, Pisani A. Loss of muscarinic autoreceptor function impairs long-term depression but not long-term potentiation in the striatum. Journal of Neuroscience. 2008;28:6258–6263. doi: 10.1523/JNEUROSCI.1678-08.2008. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

