DNA methylome reveals cellular origin of cell-free DNA in spent medium of human preimplantation embryos
- PMID: 34128477
- PMCID: PMC8203451
- DOI: 10.1172/JCI146051
DNA methylome reveals cellular origin of cell-free DNA in spent medium of human preimplantation embryos
Abstract
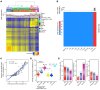
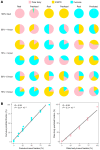
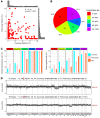
The discovery of embryonic cell-free DNA (cfDNA) in spent embryo culture media (SECM) has brought hope for noninvasive preimplantation genetic testing. However, the cellular origins of SECM cfDNA are not sufficiently understood, and methods for determining maternal DNA contamination are limited. Here, we performed whole-genome DNA methylation sequencing for SECM cfDNA. Our results demonstrated that SECM cfDNA was derived from blastocysts, cumulus cells, and polar bodies. We identified the cumulus-specific differentially methylated regions (DMRs) and oocyte/polar body-specific DMRs, and established an algorithm for deducing the cumulus, polar body, and net maternal DNA contamination ratios in SECM. We showed that DNA methylation sequencing accurately detected chromosome aneuploidy in SECM and distinguished SECM samples with low and high false negative rates and gender discordance rates, after integrating the origin analysis. Our work provides insights into the characterization of embryonic DNA in SECM and provides a perspective for noninvasive preimplantation genetic testing in reproductive medicine.
Keywords: Embryonic development; Epigenetics; Genetics; Molecular diagnosis; Reproductive Biology.
Conflict of interest statement
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

