Lactobacillus acidophilus LA14 Alleviates Liver Injury
- PMID: 34128694
- PMCID: PMC8269229
- DOI: 10.1128/mSystems.00384-21
Lactobacillus acidophilus LA14 Alleviates Liver Injury
Abstract
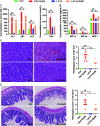
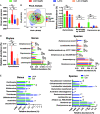
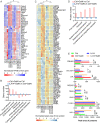
Although the probiotic Lactobacillus acidophilus LA14 is used worldwide, its effect on liver diseases remains unelucidated. Here, 32 rats were divided into four groups, gavaged with L. acidophilus LA14 (3 × 109 CFU) or phosphate-buffered saline for 7 days, and then intraperitoneally injected with d-galactosamine or saline. After 24 h, blood, liver, ileum, and feces samples were collected for liver injury, inflammation, intestinal barrier, gut microbiota, metabolome, and transcriptome analyses. Pretreatment with L. acidophilus LA14 alleviated the d-galactosamine-induced elevation of serum alanine aminotransferase (ALT), aspartate aminotransferase (AST), alkaline phosphatase (ALP), and bile acids; mitigated the histological injury to the liver and gut; and suppressed the inflammatory cytokines macrophage inflammatory protein 1α (MIP-1α), MIP-3α, and MCP-1. L. acidophilus LA14 also ameliorated the d-galactosamine-induced dysbiosis of the gut microbiota and metabolism, such as the enrichment of Bacteroides sp. strain dnLKV3 and the depletion of Streptococcus, butanoic acid, and N-acetyl-d-glucosamine. The underlying mechanism of L. acidophilus LA14 included prevention of not only the d-galactosamine-induced upregulation of infection- and tumor-related pathways but also the d-galactosamine-induced downregulation of antioxidation-related pathways during this process, as reflected by the liver transcriptome and proteome analyses. Furthermore, the administration of L. acidophilus LA14 to healthy rats did not alter the tested liver indicators but significantly enriched the beneficial Lactobacillus and Bifidobacterium species, promoted metabolism and regulated pathways to improve immunity. The ability of L. acidophilus LA14 to alleviate liver injury was further confirmed with an acetaminophen-induced mouse model. These results might provide a reference for future studies on the application of L. acidophilus LA14 for the prevention of liver injury. IMPORTANCE The probiotic Lactobacillus acidophilus LA14 is widely used, but its effect on liver diseases has not been elucidated. We explored the protective effect of L. acidophilus LA14 on the liver using rats with d-galactosamine-induced liver injury. Pretreatment with L. acidophilus LA14 alleviated the d-galactosamine-induced elevation of serum ALT, AST, ALP, and bile acids, mitigated the histological injury to the liver and gut, and suppressed the inflammatory cytokines MIP-1α, MIP-3α, and MCP-1. These effects were correlated with the modulations of the gut microbiome, metabolome, and hepatic gene expression induced by L. acidophilus LA14. Moreover, the ability of L. acidophilus LA14 to alleviate liver injury was further confirmed with an acetaminophen-induced mouse model. These results might provide a reference for future studies on the application of L. acidophilus LA14 for the prevention of liver injury.
Keywords: Lactobacillus acidophilus; acute liver injury; metabolome; microbiota; transcriptome.
Figures




Similar articles
-
Lactobacillus reuteri DSM 17938 alleviates d-galactosamine-induced liver failure in rats.Biomed Pharmacother. 2021 Jan;133:111000. doi: 10.1016/j.biopha.2020.111000. Epub 2020 Nov 14. Biomed Pharmacother. 2021. PMID: 33202285
-
Lactobacillus helveticus R0052 alleviates liver injury by modulating gut microbiome and metabolome in D-galactosamine-treated rats.Appl Microbiol Biotechnol. 2019 Dec;103(23-24):9673-9686. doi: 10.1007/s00253-019-10211-8. Epub 2019 Nov 12. Appl Microbiol Biotechnol. 2019. PMID: 31713675
-
Bifidobacterium longum R0175 Protects Rats against d-Galactosamine-Induced Acute Liver Failure.mSphere. 2020 Jan 29;5(1):e00791-19. doi: 10.1128/mSphere.00791-19. mSphere. 2020. PMID: 31996423 Free PMC article.
-
Bifidobacterium adolescentis CGMCC 15058 alleviates liver injury, enhances the intestinal barrier and modifies the gut microbiota in D-galactosamine-treated rats.Appl Microbiol Biotechnol. 2019 Jan;103(1):375-393. doi: 10.1007/s00253-018-9454-y. Epub 2018 Oct 22. Appl Microbiol Biotechnol. 2019. PMID: 30345482
-
The Mystery of Certain Lactobacillus acidophilus Strains in the Treatment of Gastrointestinal Symptoms of COVID-19: A Review.Microorganisms. 2025 Apr 19;13(4):944. doi: 10.3390/microorganisms13040944. Microorganisms. 2025. PMID: 40284780 Free PMC article. Review.
Cited by
-
Can probiotic, prebiotic, and synbiotic supplementation modulate the gut-liver axis in type 2 diabetes? A narrative and systematic review of clinical trials.Front Nutr. 2022 Dec 1;9:1052619. doi: 10.3389/fnut.2022.1052619. eCollection 2022. Front Nutr. 2022. PMID: 36532552 Free PMC article.
-
Retracted and Republished from: "Gut Microbiota Mediates the Therapeutic Effect of Monoclonal Anti-TLR4 Antibody on Acetaminophen-Induced Acute Liver Injury in Mice".Microbiol Spectr. 2023 Mar 21;11(2):e0471522. doi: 10.1128/spectrum.04715-22. Online ahead of print. Microbiol Spectr. 2023. PMID: 36942972 Free PMC article.
-
Gut Microbiota Mediates the Therapeutic Effect of Monoclonal Anti-TLR4 Antibody on Acetaminophen-Induced Acute Liver Injury in Mice.Microbiol Spectr. 2022 Jun 29;10(3):e0064722. doi: 10.1128/spectrum.00647-22. Epub 2022 May 10. Microbiol Spectr. 2022. Retraction in: Microbiol Spectr. 2023 Feb 14;11(1):e0311622. doi: 10.1128/spectrum.03116-22. PMID: 35536057 Free PMC article. Retracted.
-
Gut-Liver Axis as a Therapeutic Target for Drug-Induced Liver Injury.Curr Issues Mol Biol. 2024 Feb 1;46(2):1219-1236. doi: 10.3390/cimb46020078. Curr Issues Mol Biol. 2024. PMID: 38392196 Free PMC article. Review.
-
Vine tea extract ameliorated acute liver injury by inhibiting hepatic autophagy and reversing abnormal bile acid metabolism.Heliyon. 2023 Sep 14;9(9):e20145. doi: 10.1016/j.heliyon.2023.e20145. eCollection 2023 Sep. Heliyon. 2023. PMID: 37809393 Free PMC article.
References
-
- Milosevic I, Vujovic A, Barac A, Djelic M, Korac M, Radovanovic Spurnic A, Gmizic I, Stevanovic O, Djordjevic V, Lekic N, Russo E, Amedei A. 2019. Gut-liver axis, gut microbiota, and its modulation in the management of liver diseases: a review of the literature. Int J Mol Sci 20:395. doi:10.3390/ijms20020395. - DOI - PMC - PubMed
-
- Lv LX, Fang DQ, Shi D, Chen DY, Yan R, Zhu YX, Chen YF, Shao L, Guo FF, Wu WR, Li A, Shi HY, Jiang XW, Jiang HY, Xiao YH, Zheng SS, Li LJ. 2016. Alterations and correlations of the gut microbiome, metabolism, and immunity in patients with primary biliary cirrhosis. Environ Microbiol 18:2272–2286. doi:10.1111/1462-2920.13401. - DOI - PubMed
Grants and funding
- 11-zc03/Key Subjects of Nutrition of Zhejiang Province
- 1924D/Zhejiang Provincial Science and Technology Department
- 2018YFC2000500/MOST | National Key Research and Development Program of China (973 Program)
- 81570512/National Natural Science Foundation of China (NSFC)
- 81790631/National Natural Science Foundation of China (NSFC)
LinkOut - more resources
Full Text Sources
Miscellaneous

