Insights into evolution and coexistence of the colibactin- and yersiniabactin secondary metabolite determinants in enterobacterial populations
- PMID: 34128785
- PMCID: PMC8461471
- DOI: 10.1099/mgen.0.000577
Insights into evolution and coexistence of the colibactin- and yersiniabactin secondary metabolite determinants in enterobacterial populations
Abstract
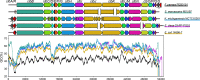
The bacterial genotoxin colibactin interferes with the eukaryotic cell cycle by causing dsDNA breaks. It has been linked to bacterially induced colorectal cancer in humans. Colibactin is encoded by a 54 kb genomic region in Enterobacteriaceae. The colibactin genes commonly co-occur with the yersiniabactin biosynthetic determinant. Investigating the prevalence and sequence diversity of the colibactin determinant and its linkage to the yersiniabactin operon in prokaryotic genomes, we discovered mainly species-specific lineages of the colibactin determinant and classified three main structural settings of the colibactin-yersiniabactin genomic region in Enterobacteriaceae. The colibactin gene cluster has a similar but not identical evolutionary track to that of the yersiniabactin operon. Both determinants could have been acquired on several occasions and/or exchanged independently between enterobacteria by horizontal gene transfer. Integrative and conjugative elements play(ed) a central role in the evolution and structural diversity of the colibactin-yersiniabactin genomic region. Addition of an activating and regulating module (clbAR) to the biosynthesis and transport module (clbB-S) represents the most recent step in the evolution of the colibactin determinant. In a first attempt to correlate colibactin expression with individual lineages of colibactin determinants and different bacterial genetic backgrounds, we compared colibactin expression of selected enterobacterial isolates in vitro. Colibactin production in the tested Klebsiella species and Citrobacter koseri strains was more homogeneous and generally higher than that in most of the Escherichia coli isolates studied. Our results improve the understanding of the diversity of colibactin determinants and its expression level, and may contribute to risk assessment of colibactin-producing enterobacteria.
Keywords: Citrobacter; Escherichia coli; Klebsiella; cytopathic effect; high pathogenicity island; polyketide; secondary metabolite.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures


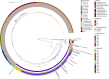



Similar articles
-
The Bacterial Stress-Responsive Hsp90 Chaperone (HtpG) Is Required for the Production of the Genotoxin Colibactin and the Siderophore Yersiniabactin in Escherichia coli.J Infect Dis. 2016 Sep 15;214(6):916-24. doi: 10.1093/infdis/jiw294. Epub 2016 Jul 13. J Infect Dis. 2016. PMID: 27412582
-
Genetic structure and distribution of the colibactin genomic island among members of the family Enterobacteriaceae.Infect Immun. 2009 Nov;77(11):4696-703. doi: 10.1128/IAI.00522-09. Epub 2009 Aug 31. Infect Immun. 2009. PMID: 19720753 Free PMC article.
-
Two Polyketides Intertwined in Complex Regulation: Posttranscriptional CsrA-Mediated Control of Colibactin and Yersiniabactin Synthesis in Escherichia coli.mBio. 2021 Feb 22;13(1):e0381421. doi: 10.1128/mbio.03814-21. Epub 2022 Feb 1. mBio. 2021. PMID: 35100864 Free PMC article.
-
Shining a Light on Colibactin Biology.Toxins (Basel). 2021 May 12;13(5):346. doi: 10.3390/toxins13050346. Toxins (Basel). 2021. PMID: 34065799 Free PMC article. Review.
-
Current understandings of colibactin regulation.Microbiology (Reading). 2024 Feb;170(2):001427. doi: 10.1099/mic.0.001427. Microbiology (Reading). 2024. PMID: 38314762 Free PMC article. Review.
Cited by
-
Dysfunctional mucus structure in cystic fibrosis increases vulnerability to colibactin-mediated DNA adducts in the colon mucosa.Gut Microbes. 2024 Jan-Dec;16(1):2387877. doi: 10.1080/19490976.2024.2387877. Epub 2024 Aug 12. Gut Microbes. 2024. PMID: 39133871 Free PMC article.
-
The Role of Cyclomodulins and Some Microbial Metabolites in Bacterial Microecology and Macroorganism Carcinogenesis.Int J Mol Sci. 2022 Oct 3;23(19):11706. doi: 10.3390/ijms231911706. Int J Mol Sci. 2022. PMID: 36233008 Free PMC article. Review.
-
Cyclomodulins and Hemolysis in E. coli as Potential Low-Cost Non-Invasive Biomarkers for Colorectal Cancer Screening.Life (Basel). 2021 Oct 31;11(11):1165. doi: 10.3390/life11111165. Life (Basel). 2021. PMID: 34833041 Free PMC article.
-
Metagenome-assembled microbial genomes from Parkinson's disease fecal samples.Sci Rep. 2024 Aug 14;14(1):18906. doi: 10.1038/s41598-024-69742-4. Sci Rep. 2024. PMID: 39143178 Free PMC article.
-
Enrichment of colibactin-associated mutational signatures in unexplained colorectal polyposis patients.BMC Cancer. 2024 Jan 18;24(1):104. doi: 10.1186/s12885-024-11849-y. BMC Cancer. 2024. PMID: 38238650 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

