A global chromatin compaction pathway that represses germline gene expression during starvation
- PMID: 34128967
- PMCID: PMC8210574
- DOI: 10.1083/jcb.202009197
A global chromatin compaction pathway that represses germline gene expression during starvation
Abstract
While much is known about how transcription is controlled at individual genes, comparatively little is known about how cells regulate gene expression on a genome-wide level. Here, we identify a molecular pathway in the C. elegans germline that controls transcription globally in response to nutritional stress. We report that when embryos hatch into L1 larvae, they sense the nutritional status of their environment, and if food is unavailable, they repress gene expression via a global chromatin compaction (GCC) pathway. GCC is triggered by the energy-sensing kinase AMPK and is mediated by a novel mechanism that involves the topoisomerase II/condensin II axis acting upstream of heterochromatin assembly. When the GCC pathway is inactivated, then transcription persists during starvation. These results define a new mode of whole-genome control of transcription.
© 2021 Belew et al.
Figures


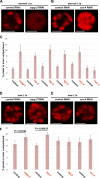

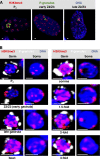
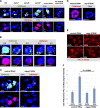


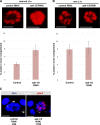

Comment in
-
Hatched and starved: Two chromatin compaction mechanisms join forces to silence germ cell genome.J Cell Biol. 2021 Sep 6;220(9):e202107026. doi: 10.1083/jcb.202107026. Epub 2021 Aug 12. J Cell Biol. 2021. PMID: 34383014 Free PMC article.

