Generation of patterned kidney organoids that recapitulate the adult kidney collecting duct system from expandable ureteric bud progenitors
- PMID: 34131121
- PMCID: PMC8206157
- DOI: 10.1038/s41467-021-23911-5
Generation of patterned kidney organoids that recapitulate the adult kidney collecting duct system from expandable ureteric bud progenitors
Abstract
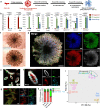
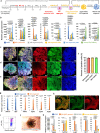
Current kidney organoids model development and diseases of the nephron but not the contiguous epithelial network of the kidney's collecting duct (CD) system. Here, we report the generation of an expandable, 3D branching ureteric bud (UB) organoid culture model that can be derived from primary UB progenitors from mouse and human fetal kidneys, or generated de novo from human pluripotent stem cells. In chemically-defined culture conditions, UB organoids generate CD organoids, with differentiated principal and intercalated cells adopting spatial assemblies reflective of the adult kidney's collecting system. Aggregating 3D-cultured nephron progenitor cells with UB organoids in vitro results in a reiterative process of branching morphogenesis and nephron induction, similar to kidney development. Applying an efficient gene editing strategy to remove RET activity, we demonstrate genetically modified UB organoids can model congenital anomalies of kidney and urinary tract. Taken together, these platforms will facilitate an enhanced understanding of development, regeneration and diseases of the mammalian collecting duct system.
Conflict of interest statement
Patent application has been filed on April 27, 2020 by University of Southern California on behalf of inventors Z.Z., B.H., A.P.M., and Z.L. for the mouse and human UB organoid generation, expansion, and CD organoid differentiation systems described in this study. The other authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

