iPLA2β-mediated lipid detoxification controls p53-driven ferroptosis independent of GPX4
- PMID: 34131139
- PMCID: PMC8206155
- DOI: 10.1038/s41467-021-23902-6
iPLA2β-mediated lipid detoxification controls p53-driven ferroptosis independent of GPX4
Abstract
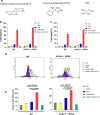
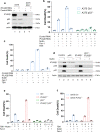
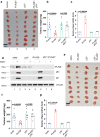
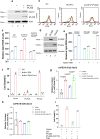
Here, we identify iPLA2β as a critical regulator for p53-driven ferroptosis upon reactive oxygen species (ROS)-induced stress. The calcium-independent phospholipase iPLA2β is known to cleave acyl tails from the glycerol backbone of lipids and release oxidized fatty acids from phospholipids. We found that iPLA2β-mediated detoxification of peroxidized lipids is sufficient to suppress p53-driven ferroptosis upon ROS-induced stress, even in GPX4-null cells. Moreover, iPLA2β is overexpressed in human cancers; inhibition of endogenous iPLA2β sensitizes tumor cells to p53-driven ferroptosis and promotes p53-dependent tumor suppression in xenograft mouse models. These results demonstrate that iPLA2β acts as a major ferroptosis repressor in a GPX4-independent manner. Notably, unlike GPX4, loss of iPLA2β has no obvious effect on normal development or cell viability in normal tissues but iPLA2β plays an essential role in regulating ferroptosis upon ROS-induced stress. Thus, our study suggests that iPLA2β is a promising therapeutic target for activating ferroptosis-mediated tumor suppression without serious toxicity concerns.
Conflict of interest statement
B.R.S. holds equity in and serves as a consultant to Inzen Therapeutics and is an inventor on patents and applications related to ferroptosis. The other authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

