Omics analyses and biochemical study of Phlebiopsis gigantea elucidate its degradation strategy of wood extractives
- PMID: 34131180
- PMCID: PMC8206109
- DOI: 10.1038/s41598-021-91756-5
Omics analyses and biochemical study of Phlebiopsis gigantea elucidate its degradation strategy of wood extractives
Abstract
Wood extractives, solvent-soluble fractions of woody biomass, are considered to be a factor impeding or excluding fungal colonization on the freshly harvested conifers. Among wood decay fungi, the basidiomycete Phlebiopsis gigantea has evolved a unique enzyme system to efficiently transform or degrade conifer extractives but little is known about the mechanism(s). In this study, to clarify the mechanism(s) of softwood degradation, we examined the transcriptome, proteome, and metabolome of P. gigantea when grown on defined media containing microcrystalline cellulose and pine sapwood extractives. Beyond the conventional enzymes often associated with cellulose, hemicellulose and lignin degradation, an array of enzymes implicated in the metabolism of softwood lipophilic extractives such as fatty and resin acids, steroids and glycerides was significantly up-regulated. Among these, a highly expressed and inducible lipase is likely responsible for lipophilic extractive degradation, based on its extracellular location and our characterization of the recombinant enzyme. Our results provide insight into physiological roles of extractives in the interaction between wood and fungi.
Conflict of interest statement
The authors declare no competing interests.
Figures
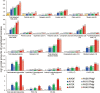
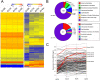




References
-
- Eriksson K, Blanchette R, Ander P. Microbial and Enzymatic Degradation of Wood and Wood Components. Springer; 1990.
-
- Rayner A, Boddy L. Fungal Decomposition of Wood: Its Biology and Ecology. Wiley; 1988.
-
- Krutul D, et al. The chemical composition of poplar wood in relation to the species and age of trees. Ann. WULS For. Wood Technol. 2019;105:125–132. doi: 10.5604/01.3001.0013.7728. - DOI
-
- Gutiérrez A, Del Río JC, González-Vila FJ, Romero J. Variation in the composition of wood extractives from Eucalyptus globulus during seasoning. J. Wood Chem. Technol. 1998;18:439–446. doi: 10.1080/02773819809349591. - DOI
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

