Advancing targeted protein degradation for cancer therapy
- PMID: 34131295
- PMCID: PMC8463487
- DOI: 10.1038/s41568-021-00365-x
Advancing targeted protein degradation for cancer therapy
Abstract
The human proteome contains approximately 20,000 proteins, and it is estimated that more than 600 of them are functionally important for various types of cancers, including nearly 400 non-enzyme proteins that are challenging to target by traditional occupancy-driven pharmacology. Recent advances in the development of small-molecule degraders, including molecular glues and heterobifunctional degraders such as proteolysis-targeting chimeras (PROTACs), have made it possible to target many proteins that were previously considered undruggable. In particular, PROTACs form a ternary complex with a hijacked E3 ubiquitin ligase and a target protein, leading to polyubiquitination and degradation of the target protein. The broad applicability of this approach is facilitated by the flexibility of individual E3 ligases to recognize different substrates. The vast majority of the approximately 600 human E3 ligases have not been explored, thus presenting enormous opportunities to develop degraders that target oncoproteins with tissue, tumour and subcellular selectivity. In this Review, we first discuss the molecular basis of targeted protein degradation. We then offer a comprehensive account of the most promising degraders in development as cancer therapies to date. Lastly, we provide an overview of opportunities and challenges in this exciting field.
© 2021. Springer Nature Limited.
Figures



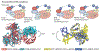

References
-
- Hart T, et al.High-resolution CRISPR screens reveal fitness genes and genotype-specific cancer liabilities. Cell 163, 1515–1526 (2015). - PubMed
-
- McDonald III ER, et al.Project DRIVE: a compendium of cancer dependencies and synthetic lethal relationships uncovered by large-scale, deep RNAi screening. Cell 170, 577–592 e10 (2017). - PubMed
-
- Behan FM, et al.Prioritization of cancer therapeutic targets using CRISPR-Cas9 screens. Nature 568, 511–516 (2019). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

