A simple one-step PCR assay for SNP detection
- PMID: 34131639
- PMCID: PMC8196326
- DOI: 10.17912/micropub.biology.000399
A simple one-step PCR assay for SNP detection
Abstract
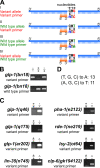
Polymerase Chain Reaction (PCR) is a powerful tool to detect natural variation or experimentally introduced variation in research and clinical settings and a widely-used method for genotyping. Single nucleotide polymorphisms (SNP) detection is challenging by PCR as the variant and wild type alleles differ by only one nucleotide. Traditional methods to detect SNPs, including Sanger sequencing and commercial kits, are usually time-consuming. Here we describe a simple primer design strategy that enables specific variant detection through regular one-step PCR. The strategy employs the differential efficiency of genomic PCR using a primer that has a single mismatch with the chromosome that contains the SNP to be detected (typically the variant allele) versus two mismatches with the corresponding alternative allele (typically the wild type allele). To date, we have successfully employed this approach to detect more than 20 SNPs. The simplicity and robustness of the approach allows rapid application to legacy mutations as well as newly discovered or generated SNPs.
Copyright: © 2021 by the authors.
Figures
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
