Overcoming chemoresistance by targeting reprogrammed metabolism: the Achilles' heel of pancreatic ductal adenocarcinoma
- PMID: 34131808
- PMCID: PMC11072422
- DOI: 10.1007/s00018-021-03866-y
Overcoming chemoresistance by targeting reprogrammed metabolism: the Achilles' heel of pancreatic ductal adenocarcinoma
Abstract
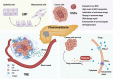
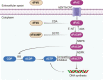
Pancreatic ductal adenocarcinoma (PDAC) is one of the leading causes of cancer-related death due to its late diagnosis that removes the opportunity for surgery and metabolic plasticity that leads to resistance to chemotherapy. Metabolic reprogramming related to glucose, lipid, and amino acid metabolism in PDAC not only enables the cancer to thrive and survive under hypovascular, nutrient-poor and hypoxic microenvironments, but also confers chemoresistance, which contributes to the poor prognosis of PDAC. In this review, we systematically elucidate the mechanism of chemotherapy resistance and the relationship of metabolic programming features with resistance to anticancer drugs in PDAC. Targeting the critical enzymes and/or transporters involved in glucose, lipid, and amino acid metabolism may be a promising approach to overcome chemoresistance in PDAC. Consequently, regulating metabolism could be used as a strategy against PDAC and could improve the prognosis of PDAC.
Keywords: Chemotherapy; Glutamine; Glycolysis; Lipogenesis; Pancreatic cancer.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

Similar articles
-
Inhibiting tumor necrosis factor-alpha diminishes desmoplasia and inflammation to overcome chemoresistance in pancreatic ductal adenocarcinoma.Oncotarget. 2016 Dec 6;7(49):81110-81122. doi: 10.18632/oncotarget.13212. Oncotarget. 2016. PMID: 27835602 Free PMC article.
-
Chemoresistance in pancreatic ductal adenocarcinoma: Overcoming resistance to therapy.Adv Cancer Res. 2023;159:285-341. doi: 10.1016/bs.acr.2023.02.010. Epub 2023 Apr 18. Adv Cancer Res. 2023. PMID: 37268399
-
Periostin promotes the chemotherapy resistance to gemcitabine in pancreatic cancer.Tumour Biol. 2016 Nov;37(11):15283-15291. doi: 10.1007/s13277-016-5321-6. Epub 2016 Sep 30. Tumour Biol. 2016. PMID: 27696296
-
Novel agents for pancreatic ductal adenocarcinoma: emerging therapeutics and future directions.J Hematol Oncol. 2018 Jan 31;11(1):14. doi: 10.1186/s13045-017-0551-7. J Hematol Oncol. 2018. PMID: 29386069 Free PMC article. Review.
-
Targeting Pancreatic Ductal Adenocarcinoma (PDAC).Cell Physiol Biochem. 2021 Jan 29;55(1):61-90. doi: 10.33594/000000326. Cell Physiol Biochem. 2021. PMID: 33508184 Review.
Cited by
-
CDKN3 Overcomes Bladder Cancer Cisplatin Resistance via LDHA-Dependent Glycolysis Reprogramming.Onco Targets Ther. 2022 Mar 26;15:299-311. doi: 10.2147/OTT.S358008. eCollection 2022. Onco Targets Ther. 2022. PMID: 35388272 Free PMC article.
-
Autophagy Contributes to Metabolic Reprogramming and Therapeutic Resistance in Pancreatic Tumors.Cells. 2022 Jan 26;11(3):426. doi: 10.3390/cells11030426. Cells. 2022. PMID: 35159234 Free PMC article. Review.
-
Extracellular vesicles derived from Lactobacillus plantarum restore chemosensitivity through the PDK2-mediated glucose metabolic pathway in 5-FU-resistant colorectal cancer cells.J Microbiol. 2022 Jul;60(7):735-745. doi: 10.1007/s12275-022-2201-1. Epub 2022 Jul 4. J Microbiol. 2022. PMID: 35781627
-
Unveiling chemotherapy-induced immune landscape remodeling and metabolic reprogramming in lung adenocarcinoma by scRNA-sequencing.Elife. 2024 Dec 27;13:RP95988. doi: 10.7554/eLife.95988. Elife. 2024. PMID: 39729352 Free PMC article.
-
AdipoRon and Pancreatic Ductal Adenocarcinoma: a future perspective in overcoming chemotherapy-induced resistance?Cancer Drug Resist. 2022 Jun 21;5(3):625-636. doi: 10.20517/cdr.2022.34. eCollection 2022. Cancer Drug Resist. 2022. PMID: 36176754 Free PMC article.
References
-
- Konstantinidis IT, Warshaw AL, Allen JN, Blaszkowsky LS, Castillo CF, Deshpande V, Hong TS, Kwak EL, Lauwers GY, Ryan DP, Wargo JA, Lillemoe KD, Ferrone CR. Pancreatic ductal adenocarcinoma: is there a survival difference for R1 resections versus locally advanced unresectable tumors? What is a "true" R0 resection? Ann Surg. 2013;257(4):731–736. doi: 10.1097/SLA.0b013e318263da2f. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

