Intrinsic OXPHOS limitations underlie cellular bioenergetics in leukemia
- PMID: 34132194
- PMCID: PMC8221809
- DOI: 10.7554/eLife.63104
Intrinsic OXPHOS limitations underlie cellular bioenergetics in leukemia
Abstract
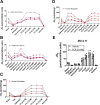
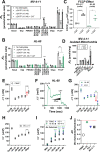
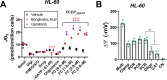
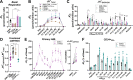
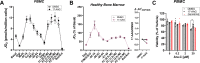
Currently there is great interest in targeting mitochondrial oxidative phosphorylation (OXPHOS) in cancer. However, notwithstanding the targeting of mutant dehydrogenases, nearly all hopeful 'mito-therapeutics' cannot discriminate cancerous from non-cancerous OXPHOS and thus suffer from a limited therapeutic index. Using acute myeloid leukemia (AML) as a model, herein, we leveraged an in-house diagnostic biochemical workflow to identify 'actionable' bioenergetic vulnerabilities intrinsic to cancerous mitochondria. Consistent with prior reports, AML growth and proliferation was associated with a hyper-metabolic phenotype which included increases in basal and maximal respiration. However, despite having nearly 2-fold more mitochondria per cell, clonally expanding hematopoietic stem cells, leukemic blasts, as well as chemoresistant AML were all consistently hallmarked by intrinsic OXPHOS limitations. Remarkably, by performing experiments across a physiological span of ATP free energy, we provide direct evidence that leukemic mitochondria are particularly poised to consume ATP. Relevant to AML biology, acute restoration of oxidative ATP synthesis proved highly cytotoxic to leukemic blasts, suggesting that active OXPHOS repression supports aggressive disease dissemination in AML. Together, these findings argue against ATP being the primary output of leukemic mitochondria and provide proof-of-principle that restoring, rather than disrupting, OXPHOS may represent an untapped therapeutic avenue for combatting hematological malignancy and chemoresistance.
Keywords: OXPHOS; acute myeloid leukemia; biochemistry; cancer bioenergetics; cancer biology; chemical biology; human; mitochondria; mitochondrial proteomics.
© 2021, Nelson et al.
Conflict of interest statement
MN, KM, JH, HC, CS, MK, KK, JM, PN, PB, NV, DL, MC, KF No competing interests declared
Figures









References
-
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society: Series B. 1995;57:289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x. - DOI
-
- Buck MD, O'Sullivan D, Klein Geltink RI, Curtis JD, Chang CH, Sanin DE, Qiu J, Kretz O, Braas D, van der Windt GJ, Chen Q, Huang SC, O'Neill CM, Edelson BT, Pearce EJ, Sesaki H, Huber TB, Rambold AS, Pearce EL. Mitochondrial dynamics controls T cell fate through metabolic programming. Cell. 2016;166:63–76. doi: 10.1016/j.cell.2016.05.035. - DOI - PMC - PubMed
-
- Byrd JC, Furman RR, Coutre SE, Flinn IW, Burger JA, Blum KA, Grant B, Sharman JP, Coleman M, Wierda WG, Jones JA, Zhao W, Heerema NA, Johnson AJ, Sukbuntherng J, Chang BY, Clow F, Hedrick E, Buggy JJ, James DF, O'Brien S. Targeting BTK with ibrutinib in relapsed chronic lymphocytic leukemia. New England Journal of Medicine. 2013;369:32–42. doi: 10.1056/NEJMoa1215637. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

