RPAcan3990: an Ultrasensitive Recombinase Polymerase Assay To Detect Angiostrongylus cantonensis DNA
- PMID: 34132583
- PMCID: PMC8373011
- DOI: 10.1128/JCM.01185-21
RPAcan3990: an Ultrasensitive Recombinase Polymerase Assay To Detect Angiostrongylus cantonensis DNA
Abstract
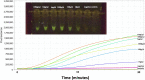
Angiostrongylus cantonensis is one of the leading causes of eosinophilic meningitis worldwide. A field-deployable molecular detection method could enhance both environmental surveillance and clinical diagnosis of this emerging pathogen. Accordingly, RPAcan3990, a recombinase polymerase assay (RPA), was developed to target a region predicted to be highly repeated in the A. cantonensis genome. The assay was then adapted to produce a visually interpretable fluorescent readout using an orange camera lens filter and a blue light. Using A. cantonensis genomic DNA, the limit of detection was found to be 1 fg/μl by both fluorometer measurement and visual reading. All clinical samples known to be positive for A. cantonensis from various areas of the globe were positive by RPAcan3990. Cerebrospinal fluid samples from other etiologies of eosinophilic meningitis (i.e., Toxocara sp. and Gnathostoma sp.) were negative in the RPAcan3990 assay. The optimal incubation temperature range for the reaction was between 35°C and 40°C. The assay successfully detected 1 fg/μl of A. cantonensis genomic DNA after incubation at human body temperature (in a shirt pocket). In conclusion, these data suggest RPAcan3990 is potentially a point-of-contact molecular assay capable of sensitively detecting A. cantonensis by producing visually interpretable results with minimal instrumentation.
Keywords: Angiostrongylus; DNA; detection; meningitis; recombinase polymerase assay.
Figures


References
-
- Stockdale Walden HD, Slapcinsky JD, Roff S, Mendieta Calle J, Diaz Goodwin Z, Stern J, Corlett R, Conway J, McIntosh A. 2017. Geographic distribution of Angiostrongylus cantonensis in wild rats (Rattus rattus) and terrestrial snails in Florida, USA. PLoS One 12:e0177910. 10.1371/journal.pone.0177910. - DOI - PMC - PubMed
-
- Guerino LR, Pecora IL, Miranda MS, Aguiar-Silva C, Carvalho OD, Caldeira RL, Silva RJ. 2017. Prevalence and distribution of Angiostrongylus cantonensis (Nematoda, Angiostrongylidae) in Achatina fulica (Mollusca, Gastropoda) in Baixada Santista, Sao Paulo, Brazil. Rev Soc Bras Med Trop 50:92–98. 10.1590/0037-8682-0316-2016. - DOI - PubMed
-
- Qvarnstrom Y, Xayavong M, da Silva AC, Park SY, Whelen AC, Calimlim PS, Sciulli RH, Honda SA, Higa K, Kitsutani P, Chea N, Heng S, Johnson S, Graeff-Teixeira C, Fox LM, da Silva AJ. 2016. Real-time polymerase chain reaction detection of Angiostrongylus cantonensis DNA in cerebrospinal fluid from patients with eosinophilic meningitis. Am J Trop Med Hyg 94:176–181. 10.4269/ajtmh.15-0146. - DOI - PMC - PubMed
-
- Sears WJ, Qvarnstrom Y, Dahlstrom E, Snook K, Kaluna L, Balaz V, Feckova B, Slapeta J, Modry D, Jarvi S, Nutman TB. 30November2020. AcanR3990 qPCR: a novel, highly sensitive, bioinformatically-informed assay to detect Angiostrongylus cantonensis infections. Clin Infect Dis 10.1093/cid/ciaa1791. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

