Detecting ALK Rearrangement with RT-PCR: A Reliable Approach Compared with Next-Generation Sequencing in Patients with NSCLC
- PMID: 34133003
- PMCID: PMC8249291
- DOI: 10.1007/s40291-021-00532-8
Detecting ALK Rearrangement with RT-PCR: A Reliable Approach Compared with Next-Generation Sequencing in Patients with NSCLC
Abstract
Background: Precise detection of anaplastic lymphoma kinase (ALK) rearrangement guides the application of ALK-targeted tyrosine kinase inhibitors (ALK-TKIs) in patients with non-small-cell lung cancer (NSCLC). Next-generation sequencing (NGS) has been widely used in clinics, but DNA-based NGS used to detect fusion genes has delivered false-negative results. However, fusion genes can be successfully detected at the transcription level and with higher sensitivity using RNA-based reverse transcription polymerase chain reaction (RT-PCR).
Objective: This study compared the performance of RT-PCR and NGS in the detection of echinoderm microtubule-associated protein-like 4 (EML4)-ALK fusion in Chinese patients with NSCLC.
Methods: Formalin-fixed paraffin-embedded tissues from 153 patients who were pathologically diagnosed as having NSCLC were collected from November 2017 to October 2019. Both DNA/RNA-based NGS and RNA-based RT-PCR were used to detect EML4-ALK fusion. For samples with discordant ALK status results, fluorescence in situ hybridization (FISH) or Sanger sequencing was used to further confirm the ALK status.
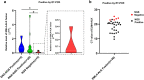
Results: In total, 124 samples were successfully analyzed using both NGS and RT-PCR. For 118 samples, results were consistent between NGS and RT-PCR, with 25 reported as ALK fusion positive and 93 as ALK fusion negative, achieving a concordance rate of 95.16%. Among the six samples with disconcordant results, five were positive using RT-PCR but negative using NGS, and one was positive using NGS but negative using RT-PCR. Four of six cases with disconcordant results (three RT-PCR positive and one NGS positive) were successfully validated using either FISH or Sanger sequencing.
Conclusions: Compared with NGS, RT-PCR appears to be a reliable method of detecting EML4-ALK fusion in patients with NSCLC.
Conflict of interest statement
Fei Yao and Changbin Zhu are employed by Amoy Diagnostics Co., Ltd. Yukun Kuang, Peihang Xu, Jiyu Wang, Yifan Zheng, Xue Sun, Zimu Li, RunJing Gan, Huixia Li, Yubiao Guo, Zunfu Ke, Kejing Tang have no conflicts of interest that are directly relevant to the content of this article.
Figures


Similar articles
-
Identification of novel ALK fusions using DNA/RNA sequencing in immunohistochemistry / RT-PCR discordant NSCLC patients.Hum Pathol. 2021 Aug;114:90-98. doi: 10.1016/j.humpath.2021.05.005. Epub 2021 May 18. Hum Pathol. 2021. PMID: 34019866
-
Comparison of IHC, FISH and RT-PCR methods for detection of ALK rearrangements in 312 non-small cell lung cancer patients in Taiwan.PLoS One. 2013 Aug 7;8(8):e70839. doi: 10.1371/journal.pone.0070839. eCollection 2013. PLoS One. 2013. PMID: 23951022 Free PMC article.
-
Comparison of reverse transcription-polymerase chain reaction, immunohistochemistry, and fluorescence in situ hybridization methodologies for detection of echinoderm microtubule-associated proteinlike 4-anaplastic lymphoma kinase fusion-positive non-small cell lung carcinoma: implications for optimal clinical testing.Arch Pathol Lab Med. 2012 Jul;136(7):796-803. doi: 10.5858/arpa.2011-0321-OA. Arch Pathol Lab Med. 2012. PMID: 22742552
-
ALK-rearrangement in non-small-cell lung cancer (NSCLC).Thorac Cancer. 2018 Apr;9(4):423-430. doi: 10.1111/1759-7714.12613. Epub 2018 Feb 28. Thorac Cancer. 2018. PMID: 29488330 Free PMC article. Review.
-
Coexistence of a novel NBEA-ALK, EML4-ALK double-fusion in a lung adenocarcinoma patient and response to alectinib: A case report.Lung Cancer. 2021 Dec;162:86-89. doi: 10.1016/j.lungcan.2021.10.015. Epub 2021 Nov 5. Lung Cancer. 2021. PMID: 34763158 Review.
Cited by
-
Targeted RNA-sequencing analysis for fusion transcripts detection in tumor diagnostics: assessment of bioinformatic tools reliability in FFPE samples.Explor Target Antitumor Ther. 2022;3(5):582-597. doi: 10.37349/etat.2022.00102. Epub 2022 Oct 27. Explor Target Antitumor Ther. 2022. PMID: 36338518 Free PMC article.
-
Current status and challenges of immunotherapy in ALK rearranged NSCLC.Front Oncol. 2022 Dec 14;12:1016869. doi: 10.3389/fonc.2022.1016869. eCollection 2022. Front Oncol. 2022. PMID: 36591504 Free PMC article.
-
Advances in the Biology, Detection Techniques, and Clinical Applications of Circulating Tumor Cells.J Oncol. 2022 Sep 2;2022:7149686. doi: 10.1155/2022/7149686. eCollection 2022. J Oncol. 2022. PMID: 36090904 Free PMC article. Review.
-
Detecting the Neuraminidase R294K Mutation in Avian Influenza A (H7N9) Virus Using Reverse Transcription Droplet Digital PCR Method.Viruses. 2023 Apr 17;15(4):983. doi: 10.3390/v15040983. Viruses. 2023. PMID: 37112963 Free PMC article.
-
A Highly Sensitive XNA-Based RT-qPCR Assay for the Identification of ALK, RET, and ROS1 Fusions in Lung Cancer.Diagnostics (Basel). 2024 Feb 24;14(5):488. doi: 10.3390/diagnostics14050488. Diagnostics (Basel). 2024. PMID: 38472960 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

