SAU-Net: A Unified Network for Cell Counting in 2D and 3D Microscopy Images
- PMID: 34133284
- PMCID: PMC8924707
- DOI: 10.1109/TCBB.2021.3089608
SAU-Net: A Unified Network for Cell Counting in 2D and 3D Microscopy Images
Abstract
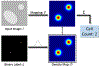
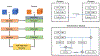
Image-based cell counting is a fundamental yet challenging task with wide applications in biological research. In this paper, we propose a novel unified deep network framework designed to solve this problem for various cell types in both 2D and 3D images. Specifically, we first propose SAU-Net for cell counting by extending the segmentation network U-Net with a Self-Attention module. Second, we design an extension of Batch Normalization (BN) to facilitate the training process for small datasets. In addition, a new 3D benchmark dataset based on the existing mouse blastocyst (MBC) dataset is developed and released to the community. Our SAU-Net achieves state-of-the-art results on four benchmark 2D datasets - synthetic fluorescence microscopy (VGG) dataset, Modified Bone Marrow (MBM) dataset, human subcutaneous adipose tissue (ADI) dataset, and Dublin Cell Counting (DCC) dataset, and the new 3D dataset, MBC. The BN extension is validated using extensive experiments on the 2D datasets, since GPU memory constraints preclude use of 3D datasets. The source code is available at https://github.com/mzlr/sau-net.
Figures









References
-
- Lempitsky V and Zisserman A, “Learning to count objects in images,” in Advances in neural information processing systems, 2010, pp. 1324–1332.
-
- Fiaschi L, Koethe U, Nair R, and Hamprecht FA, “Learning to count with regression forest and structured labels,” in Proceedings of the 21st International Conference on Pattern Recognition (ICPR2012), Nov 2012, pp. 2685–2688.

