CRISPRloci: comprehensive and accurate annotation of CRISPR-Cas systems
- PMID: 34133710
- PMCID: PMC8265192
- DOI: 10.1093/nar/gkab456
CRISPRloci: comprehensive and accurate annotation of CRISPR-Cas systems
Abstract
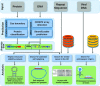
CRISPR-Cas systems are adaptive immune systems in prokaryotes, providing resistance against invading viruses and plasmids. The identification of CRISPR loci is currently a non-standardized, ambiguous process, requiring the manual combination of multiple tools, where existing tools detect only parts of the CRISPR-systems, and lack quality control, annotation and assessment capabilities of the detected CRISPR loci. Our CRISPRloci server provides the first resource for the prediction and assessment of all possible CRISPR loci. The server integrates a series of advanced Machine Learning tools within a seamless web interface featuring: (i) prediction of all CRISPR arrays in the correct orientation; (ii) definition of CRISPR leaders for each locus; and (iii) annotation of cas genes and their unambiguous classification. As a result, CRISPRloci is able to accurately determine the CRISPR array and associated information, such as: the Cas subtypes; cassette boundaries; accuracy of the repeat structure, orientation and leader sequence; virus-host interactions; self-targeting; as well as the annotation of cas genes, all of which have been missing from existing tools. This annotation is presented in an interactive interface, making it easy for scientists to gain an overview of the CRISPR system in their organism of interest. Predictions are also rendered in GFF format, enabling in-depth genome browser inspection. In summary, CRISPRloci constitutes a full suite for CRISPR-Cas system characterization that offers annotation quality previously available only after manual inspection.
© The Author(s) 2021. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Barrangou R., van der Oost J.. CRISPR–Cas Systems: RNA-mediated Adaptive Immunity in Bacteria and Archaea. 2013; Heidelburg: Springer Press; 1–129.
-
- Alkhnbashi O.S., Meier T., Mitrofanov A., Backofen R., Voss B.. CRISPR–Cas bioinformatics. Methods. 2020; 172:3–11. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

