Single-round deoxyribozyme discovery
- PMID: 34133739
- PMCID: PMC8266665
- DOI: 10.1093/nar/gkab504
Single-round deoxyribozyme discovery
Abstract
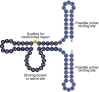
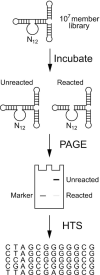
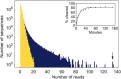
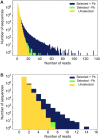
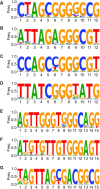
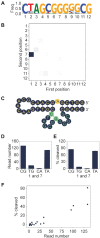
Artificial evolution experiments typically use libraries of ∼1015 sequences and require multiple rounds of selection to identify rare variants with a desired activity. Based on the simple structures of some aptamers and nucleic acid enzymes, we hypothesized that functional motifs could be isolated from significantly smaller libraries in a single round of selection followed by high-throughput sequencing. To test this idea, we investigated the catalytic potential of DNA architectures in which twelve or fifteen randomized positions were embedded in a scaffold present in all library members. After incubating in either the presence or absence of lead (which promotes the nonenzymatic cleavage of RNA), library members that cleaved themselves at an RNA linkage were purified by PAGE and characterized by high-throughput sequencing. These selections yielded deoxyribozymes with activities 8- to 30-fold lower than those previously isolated under similar conditions from libraries containing 1014 different sequences, indicating that the disadvantage of using a less diverse pool can be surprisingly small. It was also possible to elucidate the sequence requirements and secondary structures of deoxyribozymes without performing additional experiments. Due to its relative simplicity, we anticipate that this approach will accelerate the discovery of new catalytic DNA and RNA motifs.
© The Author(s) 2021. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
Similar articles
-
High-Throughput Analysis and Engineering of Ribozymes and Deoxyribozymes by Sequencing.Acc Chem Res. 2020 Dec 15;53(12):2903-2912. doi: 10.1021/acs.accounts.0c00546. Epub 2020 Nov 9. Acc Chem Res. 2020. PMID: 33164502
-
High-Throughput Activity Profiling of RNA-Cleaving DNA Catalysts by Deoxyribozyme Sequencing (DZ-seq).J Am Chem Soc. 2022 Feb 9;144(5):2090-2094. doi: 10.1021/jacs.1c12489. Epub 2022 Jan 26. J Am Chem Soc. 2022. PMID: 35081311
-
Secondary Structure Libraries for Artificial Evolution Experiments.Molecules. 2021 Mar 17;26(6):1671. doi: 10.3390/molecules26061671. Molecules. 2021. PMID: 33802780 Free PMC article.
-
Catalytic DNA: Scope, Applications, and Biochemistry of Deoxyribozymes.Trends Biochem Sci. 2016 Jul;41(7):595-609. doi: 10.1016/j.tibs.2016.04.010. Epub 2016 May 25. Trends Biochem Sci. 2016. PMID: 27236301 Free PMC article. Review.
-
The structural diversity of deoxyribozymes.Molecules. 2010 Sep 6;15(9):6269-84. doi: 10.3390/molecules15096269. Molecules. 2010. PMID: 20877222 Free PMC article. Review.
Cited by
-
Apollon: a deoxyribozyme that generates a yellow product.Nucleic Acids Res. 2024 Aug 27;52(15):9062-9075. doi: 10.1093/nar/gkae490. Nucleic Acids Res. 2024. PMID: 38869058 Free PMC article.
-
Pushing the Limits of Nucleic Acid Function.Chemistry. 2022 Dec 20;28(71):e202201737. doi: 10.1002/chem.202201737. Epub 2022 Oct 26. Chemistry. 2022. PMID: 35993619 Free PMC article. Review.
-
DNA Catalysis: Design, Function, and Optimization.Molecules. 2024 Oct 23;29(21):5011. doi: 10.3390/molecules29215011. Molecules. 2024. PMID: 39519652 Free PMC article. Review.
-
Single-Round Circular Aptamer Discovery Using Bioinspired Magnetosome-Like Magnetic Chain Cross-Linked Graphene Oxide.Research (Wash D C). 2024 May 1;7:0372. doi: 10.34133/research.0372. eCollection 2024. Research (Wash D C). 2024. PMID: 38694201 Free PMC article.
-
Aurora: a fluorescent deoxyribozyme for high-throughput screening.Nucleic Acids Res. 2024 Aug 27;52(15):9049-9061. doi: 10.1093/nar/gkae467. Nucleic Acids Res. 2024. PMID: 38860424 Free PMC article.
References
-
- Tuerk C., Gold L.. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990; 249:505–510. - PubMed
-
- Robertson D.L., Joyce G.F.. Selection in vitro of an RNA enzyme that specifically cleaves single-stranded DNA. Nature. 1990; 344:467–468. - PubMed
-
- Ellington A.D., Szostak J.W.. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990; 346:818–822. - PubMed
-
- Wilson D.S., Szostak J.W.. In vitro selection of functional nucleic acids. Annu. Rev. Biochem. 1999; 68:611–647. - PubMed
-
- Bartel D.P., Unrau P.J.. Constructing an RNA world. Trends Cell Biol. 1999; 9:M9–M13. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

