C9orf72 deficiency promotes microglial-mediated synaptic loss in aging and amyloid accumulation
- PMID: 34133945
- PMCID: PMC8298293
- DOI: 10.1016/j.neuron.2021.05.020
C9orf72 deficiency promotes microglial-mediated synaptic loss in aging and amyloid accumulation
Abstract
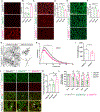
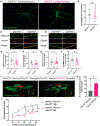
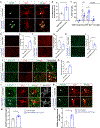
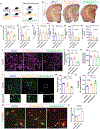
C9orf72 repeat expansions cause inherited amyotrophic lateral sclerosis (ALS)/frontotemporal dementia (FTD) and result in both loss of C9orf72 protein expression and production of potentially toxic RNA and dipeptide repeat proteins. In addition to ALS/FTD, C9orf72 repeat expansions have been reported in a broad array of neurodegenerative syndromes, including Alzheimer's disease. Here we show that C9orf72 deficiency promotes a change in the homeostatic signature in microglia and a transition to an inflammatory state characterized by an enhanced type I IFN signature. Furthermore, C9orf72-depleted microglia trigger age-dependent neuronal defects, in particular enhanced cortical synaptic pruning, leading to altered learning and memory behaviors in mice. Interestingly, C9orf72-deficient microglia promote enhanced synapse loss and neuronal deficits in a mouse model of amyloid accumulation while paradoxically improving plaque clearance. These findings suggest that altered microglial function due to decreased C9orf72 expression directly contributes to neurodegeneration in repeat expansion carriers independent of gain-of-function toxicities.
Keywords: Alzheimer’s disease; C9orf72; amyotrophic lateral sclerosis; frontotemporal dementia; microglia; neurodegeneration.
Copyright © 2021 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests R.H.B. is employed by Roche Pharmaceuticals. D.M.H. is listed as an inventor on a patent licensed by Washington University to C2N Diagnostics on the therapeutic use of anti-tau antibodies. D.M.H. co-founded and is on the scientific advisory board of C2N Diagnostics, LLC. C2N Diagnostics, LLC, has licensed certain anti-tau antibodies to AbbVie for therapeutic development. D.M.H. is on the scientific advisory board of Denali and consults for Genentech, Merck, and Cajal Neurosciences. D.M.H. and J.D.U. are listed as inventors on a provisional patent from Washington University on TREM2 antibodies. R.S. is on the scientific advisory board of Spinogenix Inc.
Figures

Comment in
-
Of mice and men: What a mouse model of microglial C9ORF72 deficiency does-and does not-tell us about human neurodegenerative diseases.Neuron. 2021 Jul 21;109(14):2203-2204. doi: 10.1016/j.neuron.2021.06.031. Neuron. 2021. PMID: 34293287 Free PMC article.
References
-
- ABO-RADY M, KALMBACH N, PAL A, SCHLUDI C, JANOSCH A, RICHTER T, FREITAG P, BICKLE M, KAHLERT AK, PETRI S, STEFANOV S, GLASS H, STAEGE S, JUST W, BHATNAGAR R, EDBAUER D, HERMANN A, WEGNER F & STERNECKERT JL 2020. Knocking out C9ORF72 Exacerbates Axonal Trafficking Defects Associated with Hexanucleotide Repeat Expansion and Reduces Levels of Heat Shock Proteins. Stem Cell Reports, 14, 390–405. - PMC - PubMed
-
- AFGAN E, BAKER D, BATUT B, VAN DEN BEEK M, BOUVIER D, CECH M, CHILTON J, CLEMENTS D, CORAOR N, GRUNING BA, GUERLER A, HILLMAN-JACKSON J, HILTEMANN S, JALILI V, RASCHE H, SORANZO N, GOECKS J, TAYLOR J, NEKRUTENKO A & BLANKENBERG D 2018. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res, 46, W537–W544. - PMC - PubMed
-
- ATANASIO A, DECMAN V, WHITE D, RAMOS M, IKIZ B, LEE HC, SIAO CJ, BRYDGES S, LAROSA E, BAI Y, FURY W, BURFEIND P, ZAMFIROVA R, WARSHAW G, ORENGO J, OYEJIDE A, FRALISH M, AUERBACH W, POUEYMIROU W, FREUDENBERG J, GONG G, ZAMBROWICZ B, VALENZUELA D, YANCOPOULOS G, MURPHY A, THURSTON G & LAI KM 2016. C9orf72 ablation causes immune dysregulation characterized by leukocyte expansion, autoantibody production, and glomerulonephropathy in mice. Sci Rep, 6, 23204. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 NS090934/NS/NINDS NIH HHS/United States
- R01 AG056303/AG/NIA NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- P30 AG066519/AG/NIA NIH HHS/United States
- P50 AG016573/AG/NIA NIH HHS/United States
- R01 AG047644/AG/NIA NIH HHS/United States
- K08 NS105916/NS/NINDS NIH HHS/United States
- RF1 AG047644/AG/NIA NIH HHS/United States
- UE5 NS065723/NS/NINDS NIH HHS/United States
- R01 AG061895/AG/NIA NIH HHS/United States
- RF1 AG055524/AG/NIA NIH HHS/United States
- RF1 NS090934/NS/NINDS NIH HHS/United States
- R01 NS097545/NS/NINDS NIH HHS/United States
- R01 NS085207/NS/NINDS NIH HHS/United States
- R25 NS065723/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

