NtbHLH1, a JAF13-like bHLH, interacts with NtMYB6 to enhance proanthocyanidin accumulation in Chinese Narcissus
- PMID: 34134615
- PMCID: PMC8207774
- DOI: 10.1186/s12870-021-03050-1
NtbHLH1, a JAF13-like bHLH, interacts with NtMYB6 to enhance proanthocyanidin accumulation in Chinese Narcissus
Abstract
Background: Flavonoid biosynthesis in plants is primarily regulated at the transcriptional level by transcription factors modulating the expression of genes encoding enzymes in the flavonoid pathway. One of the most studied transcription factor complexes involved in this regulation consists of a MYB, bHLH and WD40. However, in Chinese Narcissus (Narcissus tazetta L. var. chinensis), a popular monocot bulb flower, the regulatory mechanism of flavonoid biosynthesis remains unclear.
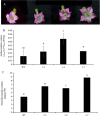
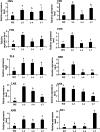
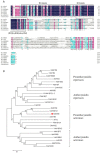
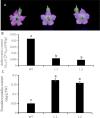
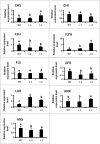
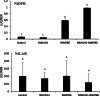
Results: In this work, genes related to the regulatory complex, NtbHLH1 and a R2R3-MYB NtMYB6, were cloned from Chinese Narcissus. Phylogenetic analysis indicated that NtbHLH1 belongs to the JAF13 clade of bHLH IIIf subgroup, while NtMYB6 was highly homologous to positive regulators of proanthocyanidin biosynthesis. Both NtbHLH1 and NtMYB6 have highest expression levels in basal plates of Narcissus, where there is an accumulation of proanthocyanidin. Ectopic over expression of NtbHLH1 in tobacco resulted in an increase in anthocyanin accumulation in flowers, and an up-regulation of expression of the endogenous tobacco bHLH AN1 and flavonoid biosynthesis genes. In contrast, the expression level of LAR gene was significantly increased in NtMYB6-transgenic tobacco. Dual luciferase assays showed that co-infiltration of NtbHLH1 and NtMYB6 significantly activated the promoter of Chinese Narcissus DFR gene. Furthermore, a yeast two-hybrid assay confirmed that NtbHLH1 interacts with NtMYB6.
Conclusions: Our results suggest that NtbHLH1 may function as a regulatory partner by interacting directly with NtMYB6 to enhance proanthocyanidin accumulation in Chinese Narcissus.
Keywords: BHLH; Chinese Narcissus; Flavonoid; MYB; Proanthocyanidin.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

