Tissue-resident macrophages provide a pro-tumorigenic niche to early NSCLC cells
- PMID: 34135508
- PMCID: PMC8923521
- DOI: 10.1038/s41586-021-03651-8
Tissue-resident macrophages provide a pro-tumorigenic niche to early NSCLC cells
Abstract
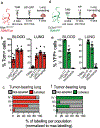
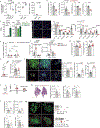
Macrophages have a key role in shaping the tumour microenvironment (TME), tumour immunity and response to immunotherapy, which makes them an important target for cancer treatment1,2. However, modulating macrophages has proved extremely difficult, as we still lack a complete understanding of the molecular and functional diversity of the tumour macrophage compartment. Macrophages arise from two distinct lineages. Tissue-resident macrophages self-renew locally, independent of adult haematopoiesis3-5, whereas short-lived monocyte-derived macrophages arise from adult haematopoietic stem cells, and accumulate mostly in inflamed lesions1. How these macrophage lineages contribute to the TME and cancer progression remains unclear. To explore the diversity of the macrophage compartment in human non-small cell lung carcinoma (NSCLC) lesions, here we performed single-cell RNA sequencing of tumour-associated leukocytes. We identified distinct populations of macrophages that were enriched in human and mouse lung tumours. Using lineage tracing, we discovered that these macrophage populations differ in origin and have a distinct temporal and spatial distribution in the TME. Tissue-resident macrophages accumulate close to tumour cells early during tumour formation to promote epithelial-mesenchymal transition and invasiveness in tumour cells, and they also induce a potent regulatory T cell response that protects tumour cells from adaptive immunity. Depletion of tissue-resident macrophages reduced the numbers and altered the phenotype of regulatory T cells, promoted the accumulation of CD8+ T cells and reduced tumour invasiveness and growth. During tumour growth, tissue-resident macrophages became redistributed at the periphery of the TME, which becomes dominated by monocyte-derived macrophages in both mouse and human NSCLC. This study identifies the contribution of tissue-resident macrophages to early lung cancer and establishes them as a target for the prevention and treatment of early lung cancer lesions.
© 2021. The Author(s), under exclusive licence to Springer Nature Limited.
Figures






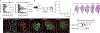
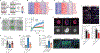
Comment in
-
Home advantage for tissue-resident macrophages.Nat Rev Cancer. 2021 Sep;21(9):539. doi: 10.1038/s41568-021-00393-7. Nat Rev Cancer. 2021. PMID: 34294891 No abstract available.
-
Targeting resident macrophages in cancer.Nat Immunol. 2021 Sep;22(9):1078-1079. doi: 10.1038/s41590-021-01002-3. Nat Immunol. 2021. PMID: 34354280 Free PMC article. No abstract available.
References
-
- Schulz C et al. A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science 336, 86–90 (2012). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

