Host-Parasite Interaction of Atlantic salmon (Salmo salar) and the Ectoparasite Neoparamoeba perurans in Amoebic Gill Disease
- PMID: 34135900
- PMCID: PMC8202022
- DOI: 10.3389/fimmu.2021.672700
Host-Parasite Interaction of Atlantic salmon (Salmo salar) and the Ectoparasite Neoparamoeba perurans in Amoebic Gill Disease
Abstract
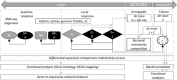
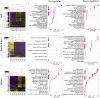
Marine farmed Atlantic salmon (Salmo salar) are susceptible to recurrent amoebic gill disease (AGD) caused by the ectoparasite Neoparamoeba perurans over the growout production cycle. The parasite elicits a highly localized response within the gill epithelium resulting in multifocal mucoid patches at the site of parasite attachment. This host-parasite response drives a complex immune reaction, which remains poorly understood. To generate a model for host-parasite interaction during pathogenesis of AGD in Atlantic salmon the local (gill) and systemic transcriptomic response in the host, and the parasite during AGD pathogenesis was explored. A dual RNA-seq approach together with differential gene expression and system-wide statistical analyses of gene and transcription factor networks was employed. A multi-tissue transcriptomic data set was generated from the gill (including both lesioned and non-lesioned tissue), head kidney and spleen tissues naïve and AGD-affected Atlantic salmon sourced from an in vivo AGD challenge trial. Differential gene expression of the salmon host indicates local and systemic upregulation of defense and immune responses. Two transcription factors, znfOZF-like and znf70-like, and their associated gene networks significantly altered with disease state. The majority of genes in these networks are candidates for mediators of the immune response, cellular proliferation and invasion. These include Aurora kinase B-like, rho guanine nucleotide exchange factor 25-like and protein NDNF-like inhibited. Analysis of the N. perurans transcriptome during AGD pathology compared to in vitro cultured N. perurans trophozoites, as a proxy for wild type trophozoites, identified multiple gene candidates for virulence and indicates a potential master regulatory gene system analogous to the two-component PhoP/Q system. Candidate genes identified are associated with invasion of host tissue, evasion of host defense mechanisms and formation of the mucoid lesion. We generated a novel model for host-parasite interaction during AGD pathogenesis through integration of host and parasite functional profiles. Collectively, this dual transcriptomic study provides novel molecular insights into the pathology of AGD and provides alternative theories for future research in a step towards improved management of AGD.
Keywords: Atlantic salmon (Salmo salar); Neoparamoeba perurans; amoebic gill disease; aquaculture; dual RNA-Seq; host-parasite interaction; immunity.
Copyright © 2021 Botwright, Mohamed, Slinger, Lima and Wynne.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Munday BL, Zilberg D, Findlay V. Gill Disease of Marine Fish Caused by Infection With Neoparamoeba Pemaquidensis . J Fish Dis (2001) 24:497–507. 10.1046/j.1365-2761.2001.00329.x - DOI
-
- Powell MD, Reynolds P, Kristensen T. Freshwater Treatment of Amoebic Gill Disease and Sea-Lice in Seawater Salmon Production: Considerations of Water Chemistry and Fish Welfare in Norway. Aquaculture (2015) 448:18–28. 10.1016/j.aquaculture.2015.05.027 - DOI
-
- Kube PD, Taylor RS, Elliott NG. Genetic Variation in Parasite Resistance of Atlantic Salmon to Amoebic Gill Disease Over Multiple Infections. Aquaculture (2012) 364–365:165–72. 10.1016/j.aquaculture.2012.08.026 - DOI
Publication types
MeSH terms
Supplementary concepts
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

