The global emergence of a novel Streptococcus suis clade associated with human infections
- PMID: 34137500
- PMCID: PMC8261479
- DOI: 10.15252/emmm.202013810
The global emergence of a novel Streptococcus suis clade associated with human infections
Abstract
Streptococcus suis, a ubiquitous bacterial colonizer in pigs, has recently extended host range to humans, leading to a global surge of deadly human infections and three large outbreaks since 1998. To better understand the mechanisms for the emergence of cross-species transmission and virulence in human, we have sequenced 366 S. suis human and pig isolates from 2005 to 2016 and performed a large-scale phylogenomic analysis on 1,634 isolates from 14 countries over 36 years. We show the formation of a novel human-associated clade (HAC) diversified from swine S. suis isolates. Phylogeographic analysis identified Europe as the origin of HAC, coinciding with the exportation of European swine breeds between 1960s and 1970s. HAC is composed of three sub-lineages and contains several healthy-pig isolates that display high virulence in experimental infections, suggesting healthy-pig carriers as a potential source for human infection. New HAC-specific genes are identified as promising markers for pathogen detection and surveillance. Our discovery of a human-associated S. suis clade provides insights into the evolution of this emerging human pathogen and extend our understanding of S. suis epidemics worldwide.
Keywords: Streptococcus suis; ST1; ST7; human pathogen; population genomics.
© 2021 The Authors. Published under the terms of the CC BY 4.0 license.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

Correlation of the maximum‐likelihood tree with genetic and phenotypic classification/clustering. Maximum‐likelihood tree was constructed using genome‐wide SNPs of 1,634 S. suis isolates. Tip nodes are colored based on the host source (human in red; diseased pig in yellow; and healthy pig in green). From inside to outside: The first ring represents BAPS clustering; the second ring represents the country source. Human‐associated clade (HAC) are colored in red, with diseased‐pig clade (DPC) in yellow and healthy‐pig clade (HPC) in green. Phylogenetic relationship is consistent with BAPS clustering and correlates with host source. Most isolates in BAPS2, BAPS5, and BAPS6 were obtained from healthy pigs. The majority of isolates in BAPS7 and BAPS8 were obtained from patients and diseased pigs, respectively.
Phylogenetic tree (unrooted) showing three important clades (HAC in red; DPC in yellow; and HPC in green).

The violin plot depicting the smaller genome size of HAC isolates (n = 820) than that of DPC (n = 407) and HPC (n = 262). Inside the violins are the box and whisker plots, with the dot showing the median, the box showing the quartiles, and the whisker showing the 95% percentiles. P‐values were calculated using unpaired t‐test (**P < 0.05).
The survival curve for zebrafish inoculated with representative S. suis isolates from HAC and HPC.

Bayesian skyline plot showing the population size through time for human‐associated S. suis, which is constructed based on a subsample of HAC isolates (n = 174). The y‐axis represents the effective population size, and the x‐axis is calendar years. The line shows the median estimate of the population size. Blue shading shows 95% highest posterior density.
Timed phylogeny of the subsample of HAC isolates (n = 174). Maximum clade credibility tree is produced using strict‐clock model in BEAST2. Major sub‐lineages in Asia are indicated (I, II, and III). Acquisition of the 89‐kb pathogenicity island is indicated by a red pentagon. Acquisition of the 78‐kb pathogenicity island and the 127‐kb MGE is indicated by black and green pentagon, respectively. The blue and red blocks of heatmaps represent the presence of MGEs and ARGs, respectively. Gray represents absence.
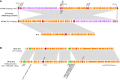
Two types of 127‐kb tandem MGE detected in multi‐drug‐resistant S. suis ST7 isolates in lineage III. The 127‐kb MGE encodes eight antibiotic resistance genes. The integrative and conjugative elements (ICE, in orange) and prophage (in purple), both of which are flanked by att sites, are tandemly arranged in two possible orders. The antibiotic resistance genes are indicated in red. The chromosomal conserved genes around MGE are indicated in black.
Comparison of the novel 78‐kb PAI reported in this study, with the 89‐kb PAI reported in epidemic strains from two outbreaks in China. The key factors for virulence of epidemic strains are indicated in green, including SalKR, NisKR, a type IV‐like secretion system, and a Tn916 element. The antibiotic resistance genes are shown in red. Three regions in the 89‐kb PAI are absent from the 78‐kb PAI. No virulence‐related genes were found within these regions.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

