Human STAT3 variants underlie autosomal dominant hyper-IgE syndrome by negative dominance
- PMID: 34137790
- PMCID: PMC8217968
- DOI: 10.1084/jem.20202592
Human STAT3 variants underlie autosomal dominant hyper-IgE syndrome by negative dominance
Abstract
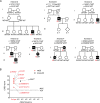
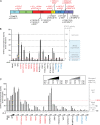
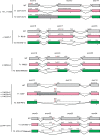
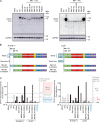
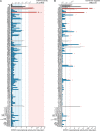
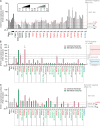
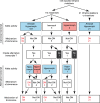
Most patients with autosomal dominant hyper-IgE syndrome (AD-HIES) carry rare heterozygous STAT3 variants. Only six of the 135 in-frame variants reported have been experimentally shown to be dominant negative (DN), and it has been recently suggested that eight out-of-frame variants operate by haploinsufficiency. We experimentally tested these 143 variants, 7 novel out-of-frame variants found in HIES patients, and other STAT3 variants from the general population. Strikingly, all 15 out-of-frame variants were DN via their encoded (1) truncated proteins, (2) neoproteins generated from a translation reinitiation codon, and (3) isoforms from alternative transcripts or a combination thereof. Moreover, 128 of the 135 in-frame variants (95%) were also DN. The patients carrying the seven non-DN STAT3 in-frame variants have not been studied for other genetic etiologies. Finally, none of the variants from the general population tested, including an out-of-frame variant, were DN. Overall, our findings show that heterozygous STAT3 variants, whether in or out of frame, underlie AD-HIES through negative dominance rather than haploinsufficiency.
© 2021 Asano et al.
Conflict of interest statement
Disclosures: The authors declare no competing interests exist.
Figures







References
-
- Abolhassani, H., Chou J., Bainter W., Platt C.D., Tavassoli M., Momen T., Tavakol M., Eslamian M.H., Gharagozlou M., Movahedi M., et al. . 2018. Clinical, immunologic, and genetic spectrum of 696 patients with combined immunodeficiency. J. Allergy Clin. Immunol. 141:1450–1458. 10.1016/j.jaci.2017.06.049 - DOI - PubMed
-
- Al Khatib, S., Keles S., Garcia-Lloret M., Karakoc-Aydiner E., Reisli I., Artac H., Camcioglu Y., Cokugras H., Somer A., Kutukculer N., et al. . 2009. Defects along the T(H)17 differentiation pathway underlie genetically distinct forms of the hyper IgE syndrome. J. Allergy Clin. Immunol. 124:342–348: 348.e1–348.e5. 10.1016/j.jaci.2009.05.004 - DOI - PMC - PubMed
-
- Alcántara-Montiel, J.C., Staines-Boone T., López-Herrera G., Espinosa-Rosales F., Espinosa-Padilla S.E., Hernández-Rivas R., and Santos-Argumedo L.. 2016. Functional characterization of two new STAT3 mutations associated with hyper-IgE syndrome in a Mexican cohort. Clin. Genet. 89:217–221. 10.1111/cge.12658 - DOI - PubMed

