Insights from Binding on Quadruplex Selective Carbazole Ligands
- PMID: 34138492
- PMCID: PMC8518889
- DOI: 10.1002/chem.202101866
Insights from Binding on Quadruplex Selective Carbazole Ligands
Abstract
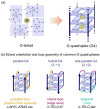
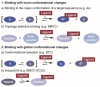
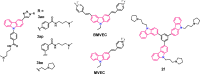
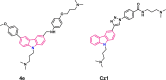
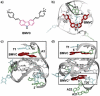
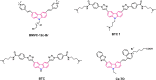
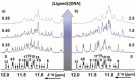
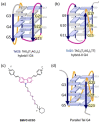
Polymorphic G-quadruplex (G4) secondary DNA structures have received increasing attention in medicinal chemistry owing to their key involvement in the regulation of the maintenance of genomic stability, telomere length homeostasis and transcription of important proto-oncogenes. Different classes of G4 ligands have been developed for the potential treatment of several human diseases. Among them, the carbazole scaffold with appropriate side chain appendages has attracted much interest for designing G4 ligands. Because of its large and rigid π-conjugation system and ease of functionalization at three different positions, a variety of carbazole derivatives have been synthesized from various natural or synthetic sources for potential applications in G4-based therapeutics and biosensors. Herein, we provide an updated close-up of the literatures on carbazole-based G4 ligands with particular focus given on their detailed binding insights studied by NMR spectroscopy. The structure-activity relationships and the opportunities and challenges of their potential applications as biosensors and therapeutics are also discussed. This review will provide an overall picture of carbazole ligands with remarkable G4 topological preference, fluorescence properties and significant bioactivity; portraying carbazole as a very promising scaffold for assembling G4 ligands with a range of novel functional applications.
Keywords: G-quadruplexes; NMR spectroscopy; biophysics; carbazole ligands; ligand design.
© 2021 The Authors. Chemistry - A European Journal published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- None
-
- Han H., Hurley L. H., Trends Pharmacol. Sci. 2000, 21, 136–142; - PubMed
-
- Amrane S., Kerkour A., Bedrat A., Vialet B., Andreola M. L., Mergny J. L., J. Am. Chem. Soc. 2014, 136, 5249–5252; - PubMed
-
- Haensel-Hertsch R., Simeone A., Shea A., Hui W. W. I., Zyner K. G., Marsico G., Rueda O. M., Bruna A., Martin A., Zhang X., Adhikari S., Tannahill D., Caldas C., Balasubramanian S., Nat. Genet. 2020, 52, 878–883; - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

