Comparative evaluation of isogenic mesodermal and ectomesodermal chondrocytes from human iPSCs for cartilage regeneration
- PMID: 34138734
- PMCID: PMC8133756
- DOI: 10.1126/sciadv.abf0907
Comparative evaluation of isogenic mesodermal and ectomesodermal chondrocytes from human iPSCs for cartilage regeneration
Abstract
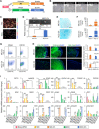
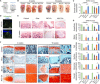
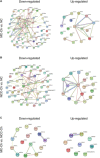
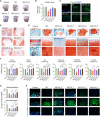
Generating phenotypic chondrocytes from pluripotent stem cells is of great interest in the field of cartilage regeneration. In this study, we differentiated human induced pluripotent stem cells into the mesodermal and ectomesodermal lineages to prepare isogenic mesodermal cell-derived chondrocytes (MC-Chs) and neural crest cell-derived chondrocytes (NCC-Chs), respectively, for comparative evaluation. Our results showed that both MC-Chs and NCC-Chs expressed hyaline cartilage-associated markers and were capable of generating hyaline cartilage-like tissue ectopically and at joint defects. Moreover, NCC-Chs revealed closer morphological and transcriptional similarities to native articular chondrocytes than MC-Chs. NCC-Ch implants induced by our growth factor mixture demonstrated increased matrix production and stiffness compared to MC-Ch implants. Our findings address how chondrocytes derived from pluripotent stem cells through mesodermal and ectomesodermal differentiation are different in activities and functions, providing the crucial information that helps make appropriate cell choices for effective regeneration of articular cartilage.
Copyright © 2021 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works. Distributed under a Creative Commons Attribution NonCommercial License 4.0 (CC BY-NC).
Figures




References
-
- B. M. Carlson, Human Embryology and Developmental Biology E-Book (Elsevier Health Sciences, 2018).
-
- P. Trainor, Neural Crest Cells: Evolution, Development and Disease (Academic Press, 2013).
-
- U.S. National Library of Medicine “ClinicalTrials.gov” National Institutes of Health (2021); www.clinicaltrials.gov/ct2/home.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

