Mechanical diversity and folding intermediates of parallel-stranded G-quadruplexes with a bulge
- PMID: 34139007
- PMCID: PMC8266575
- DOI: 10.1093/nar/gkab531
Mechanical diversity and folding intermediates of parallel-stranded G-quadruplexes with a bulge
Abstract
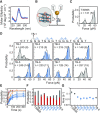
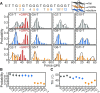
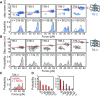
A significant number of sequences in the human genome form noncanonical G-quadruplexes (G4s) with bulges or a guanine vacancy. Here, we systematically characterized the mechanical stability of parallel-stranded G4s with a one to seven nucleotides bulge at various positions. Our results show that G4-forming sequences with a bulge form multiple conformations, including fully-folded G4 with high mechanical stability (unfolding forces > 40 pN), partially-folded intermediates (unfolding forces < 40 pN). The folding probability and folded populations strongly depend on the positions and lengths of the bulge. By combining a single-molecule unfolding assay, dimethyl sulfate (DMS) footprinting, and a guanine-peptide conjugate that selectively stabilizes guanine-vacancy-bearing G-quadruplexes (GVBQs), we identified that GVBQs are the major intermediates of G4s with a bulge near the 5' or 3' ends. The existence of multiple structures may induce different regulatory functions in many biological processes. This study also demonstrates a new strategy for selectively stabilizing the intermediates of bulged G4s to modulate their functions.
© The Author(s) 2021. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Sen D., Gilbert W.. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature. 1988; 334:364–366. - PubMed
-
- Davis J.T. G-quartets 40 years later: from 5′-GMP to molecular biology and supramolecular chemistry. Angew. Chem. Int. Ed. 2004; 43:668–698. - PubMed
-
- Hansel-Hertsch R., Di Antonio M., Balasubramanian S.. DNA G-quadruplexes in the human genome: detection, functions and therapeutic potential. Nat. Rev. Mol. Cell Biol. 2017; 18:279–284. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

