DnaB helicase dynamics in bacterial DNA replication resolved by single-molecule studies
- PMID: 34139009
- PMCID: PMC8266626
- DOI: 10.1093/nar/gkab493
DnaB helicase dynamics in bacterial DNA replication resolved by single-molecule studies
Abstract
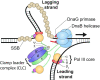
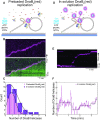
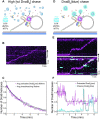
In Escherichia coli, the DnaB helicase forms the basis for the assembly of the DNA replication complex. The stability of DnaB at the replication fork is likely important for successful replication initiation and progression. Single-molecule experiments have significantly changed the classical model of highly stable replication machines by showing that components exchange with free molecules from the environment. However, due to technical limitations, accurate assessments of DnaB stability in the context of replication are lacking. Using in vitro fluorescence single-molecule imaging, we visualise DnaB loaded on forked DNA templates. That these helicases are highly stable at replication forks, indicated by their observed dwell time of ∼30 min. Addition of the remaining replication factors results in a single DnaB helicase integrated as part of an active replisome. In contrast to the dynamic behaviour of other replisome components, DnaB is maintained within the replisome for the entirety of the replication process. Interestingly, we observe a transient interaction of additional helicases with the replication fork. This interaction is dependent on the τ subunit of the clamp-loader complex. Collectively, our single-molecule observations solidify the role of the DnaB helicase as the stable anchor of the replisome, but also reveal its capacity for dynamic interactions.
© The Author(s) 2021. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



Similar articles
-
A Primase-Induced Conformational Switch Controls the Stability of the Bacterial Replisome.Mol Cell. 2020 Jul 2;79(1):140-154.e7. doi: 10.1016/j.molcel.2020.04.037. Epub 2020 May 27. Mol Cell. 2020. PMID: 32464091 Free PMC article.
-
Overexpression of the Replicative Helicase in Escherichia coli Inhibits Replication Initiation and Replication Fork Reloading.J Mol Biol. 2016 Mar 27;428(6):1068-1079. doi: 10.1016/j.jmb.2016.01.018. Epub 2016 Jan 23. J Mol Biol. 2016. PMID: 26812209 Free PMC article.
-
Localization of an accessory helicase at the replisome is critical in sustaining efficient genome duplication.Nucleic Acids Res. 2011 Feb;39(3):949-57. doi: 10.1093/nar/gkq889. Epub 2010 Oct 4. Nucleic Acids Res. 2011. PMID: 20923786 Free PMC article.
-
Whither the replisome: emerging perspectives on the dynamic nature of the DNA replication machinery.Cell Cycle. 2009 Sep 1;8(17):2686-91. doi: 10.4161/cc.8.17.9390. Epub 2009 Sep 29. Cell Cycle. 2009. PMID: 19652539 Free PMC article. Review.
-
Structural Insight Into the Function of DnaB Helicase in Bacterial DNA Replication.Proteins. 2025 Feb;93(2):420-429. doi: 10.1002/prot.26746. Epub 2024 Sep 4. Proteins. 2025. PMID: 39230358 Review.
Cited by
-
Single-molecule visualization of stalled replication-fork rescue by the Escherichia coli Rep helicase.Nucleic Acids Res. 2023 Apr 24;51(7):3307-3326. doi: 10.1093/nar/gkad186. Nucleic Acids Res. 2023. PMID: 36938885 Free PMC article.
-
A bipartite interaction with the processivity clamp potentiates Pol IV-mediated TLS.bioRxiv [Preprint]. 2024 May 31:2024.05.30.596738. doi: 10.1101/2024.05.30.596738. bioRxiv. 2024. Update in: Proc Natl Acad Sci U S A. 2025 Mar 04;122(9):e2421471122. doi: 10.1073/pnas.2421471122. PMID: 38853898 Free PMC article. Updated. Preprint.
-
Insight into Single-Molecule Imaging Techniques for the Study of Prokaryotic Genome Maintenance.Chem Biomed Imaging. 2024 Jun 18;2(9):595-614. doi: 10.1021/cbmi.4c00037. eCollection 2024 Sep 23. Chem Biomed Imaging. 2024. PMID: 39328428 Free PMC article. Review.
-
The Caulobacter crescentus DciA promotes chromosome replication through topological loading of the DnaB replicative helicase at replication forks.Nucleic Acids Res. 2022 Dec 9;50(22):12896-12912. doi: 10.1093/nar/gkac1146. Nucleic Acids Res. 2022. PMID: 36484102 Free PMC article.
-
Dysregulated DnaB unwinding induces replisome decoupling and daughter strand gaps that are countered by RecA polymerization.Nucleic Acids Res. 2024 Jul 8;52(12):6977-6993. doi: 10.1093/nar/gkae435. Nucleic Acids Res. 2024. PMID: 38808668 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

