Transcriptomic analysis of zebrafish prion protein mutants supports conserved cross-species function of the cellular prion protein
- PMID: 34139950
- PMCID: PMC8216189
- DOI: 10.1080/19336896.2021.1924557
Transcriptomic analysis of zebrafish prion protein mutants supports conserved cross-species function of the cellular prion protein
Abstract
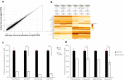
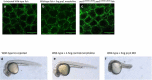
Cellular Prion Protein (PrPC) is a well-studied protein as the substrate for various progressive untreatable neurodegenerative diseases. Normal functions of PrPC are poorly understood, though recent proteomic and transcriptomic approaches have begun to reveal common themes. We use our compound prp1 and prp2 knockout mutant zebrafish at three days post fertilization to take a transcriptomic approach to investigating potentially conserved PrPC functions during development. Gene ontology analysis shows the biological processes with the largest changes in gene expression include redox processing, transport and cell adhesion. Within these categories several different gene families were prevalent including the solute carrier proteins, cytochrome p450 enzymes and protocadherins. Continuing from previous studies identifying cell adhesion as an important function of PrPC we found that in addition to the protocadherins there was a significant reduction in transcript abundance of both ncam1a and st8sia2. These two genes are involved in the early development of vertebrates. The alterations in cell adhesion transcripts were consistent with past findings in zebrafish and mouse prion protein mutants; however E-cadherin processing after prion protein knockdown failed to reveal any differences compared with wild type in either our double prp1/prp2 mutant fish or after prp1 morpholino knockdown. Our data supports a cross species conserved role for PrPC in the development and maintenance of the central nervous system, particularly by regulating various and important cell adhesion processes.
Keywords: Prion knockout; RNA-sequencing; cell adhesion; scrapie; transcriptome.
Conflict of interest statement
The authors have no competing interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
