Rapid, simplified whole blood-based multiparameter assay to quantify and phenotype SARS-CoV-2-specific T-cells
- PMID: 34140294
- PMCID: PMC8215505
- DOI: 10.1183/13993003.00285-2021
Rapid, simplified whole blood-based multiparameter assay to quantify and phenotype SARS-CoV-2-specific T-cells
Abstract
Background: Rapid tests to evaluate severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)-specific T-cell responses are urgently needed to decipher protective immunity and aid monitoring vaccine-induced immunity.
Methods: Using a rapid whole blood assay requiring a minimal amount of blood, we measured qualitatively and quantitatively SARS-CoV-2-specific CD4 T-cell responses in 31 healthcare workers using flow cytometry.
Results: 100% of COVID-19 convalescent participants displayed a detectable SARS-CoV-2-specific CD4 T-cell response. SARS-CoV-2-responding cells were also detected in 40.9% of participants with no COVID-19-associated symptoms or who tested PCR-negative. Phenotypic assessment indicated that, in COVID-19 convalescent participants, SARS-CoV-2 CD4 responses displayed an early differentiated memory phenotype with limited capacity to produce interferon (IFN)-γ. Conversely, in participants with no reported symptoms, SARS-CoV-2 CD4 responses were enriched in late differentiated cells, coexpressing IFN-γ and tumour necrosis factor-α and also Granzyme B.
Conclusions: This proof-of-concept study presents a scalable alternative to peripheral blood mononuclear cell-based assays to enumerate and phenotype SARS-CoV-2-responding T-cells, thus representing a practical tool to monitor adaptive immunity due to natural infection or vaccine trials.
Copyright ©The authors 2022.
Conflict of interest statement
Conflict of interest: C. Riou has nothing to disclose. Conflict of interest: G. Schäfer has nothing to disclose. Conflict of interest: E. du Bruyn has nothing to disclose. Conflict of interest: R.T. Goliath has nothing to disclose. Conflict of interest: C. Stek has nothing to disclose. Conflict of interest: H. Mou has nothing to disclose. Conflict of interest: D. Hung has nothing to disclose. Conflict of interest: K.A. Wilkinson has nothing to disclose. Conflict of interest: R.J. Wilkinson reports grants from Wellcome, Cancer Research UK, UK Research and Innovation, and the European and Developing Countries Clinical Trials Partnership, during the conduct of the study.
Figures
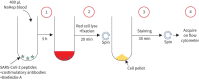


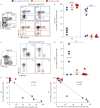

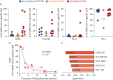
Update of
-
Rapid, simplified whole blood-based multiparameter assay to quantify and phenotype SARS-CoV-2 specific T cells.medRxiv [Preprint]. 2020 Nov 3:2020.10.30.20223099. doi: 10.1101/2020.10.30.20223099. medRxiv. 2020. Update in: Eur Respir J. 2022 Jan 13;59(1):2100285. doi: 10.1183/13993003.00285-2021. PMID: 33173918 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous
