Epitranscriptomic addition of m6A regulates HIV-1 RNA stability and alternative splicing
- PMID: 34140354
- PMCID: PMC8247604
- DOI: 10.1101/gad.348508.121
Epitranscriptomic addition of m6A regulates HIV-1 RNA stability and alternative splicing
Abstract
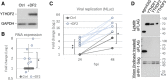
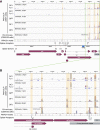
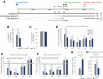
Previous work has demonstrated that the epitranscriptomic addition of m6A to viral transcripts can promote the replication and pathogenicity of a wide range of DNA and RNA viruses, including HIV-1, yet the underlying mechanisms responsible for this effect have remained unclear. It is known that m6A function is largely mediated by cellular m6A binding proteins or readers, yet how these regulate viral gene expression in general, and HIV-1 gene expression in particular, has been controversial. Here, we confirm that m6A addition indeed regulates HIV-1 RNA expression and demonstrate that this effect is largely mediated by the nuclear m6A reader YTHDC1 and the cytoplasmic m6A reader YTHDF2. Both YTHDC1 and YTHDF2 bind to multiple distinct and overlapping sites on the HIV-1 RNA genome, with YTHDC1 recruitment serving to regulate the alternative splicing of HIV-1 RNAs. Unexpectedly, while YTHDF2 binding to m6A residues present on cellular mRNAs resulted in their destabilization as previously reported, YTHDF2 binding to m6A sites on HIV-1 transcripts resulted in a marked increase in the stability of these viral RNAs. Thus, YTHDF2 binding can exert diametrically opposite effects on RNA stability, depending on RNA sequence context.
Keywords: HIV-1; RNA; RNA stability; YTHDC1; YTHDF2; epitranscriptomic regulation; splicing.
© 2021 Tsai et al.; Published by Cold Spring Harbor Laboratory Press.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
