Expression of a maize SOC1 gene enhances soybean yield potential through modulating plant growth and flowering
- PMID: 34140602
- PMCID: PMC8211702
- DOI: 10.1038/s41598-021-92215-x
Expression of a maize SOC1 gene enhances soybean yield potential through modulating plant growth and flowering
Abstract
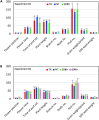
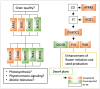
Yield enhancement is a top priority for soybean (Glycine max Merr.) breeding. SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1) is a major integrator in flowering pathway, and it is anticipated to be capable of regulating soybean reproductive stages through its interactions with other MADS-box genes. Thus, we produced transgenic soybean for a constitutive expression of a maize SOC1 (ZmSOC1). T1 transgenic plants, in comparison with the nontransgenic plants, showed early flowering, reduced height of mature plants, and no significant impact on grain quality. The transgenic plants also had a 13.5-23.2% of higher grain weight per plant than the nontransgenic plants in two experiments. Transcriptome analysis in the leaves of 34-day old plants revealed 58 differentially expressed genes (DEGs) responding to the expression of the ZmSOC1, of which the upregulated FRUITFULL MADS-box gene, as well as the transcription factor VASCULAR PLANT ONE-ZINC FINGER1, contributed to the promoted flowering. The downregulated gibberellin receptor GID1B could play a major role in reducing the plant height. The remaining DEGs suggested broader effects on the other unmeasured traits (e.g., photosynthesis efficiency and abiotic tolerance), which could contribute to yield increase. Overall, modulating expression of SOC1 in soybean provides a novel and promising approach to regulate plant growth and reproductive development and thus has a potential either to enhance grain yield or to change plant adaptability.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Pastor AV, et al. The global nexus of food-trade-water sustaining environmental flows by 2050. Nat. Sustain. 2019;2:499–507. doi: 10.1038/s41893-019-0287-1. - DOI
-
- Bhattarai MD, Secchi S, Schoof J. Projecting corn and soybeans yields under climate change in a Corn Belt watershed. Agric. Syst. 2017;152:90–99. doi: 10.1016/j.agsy.2016.12.013. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

